predict
Description
yfit = predict(Mdl,X,Name=Value)
Examples
Predict test set response values by using a trained regression neural network model.
Load the patients data set. Create a table from the data set. Each row corresponds to one patient, and each column corresponds to a diagnostic variable. Use the Systolic variable as the response variable, and the rest of the variables as predictors.
load patients
tbl = table(Diastolic,Height,Smoker,Weight,Systolic);Separate the data into a training set tblTrain and a test set tblTest by using a nonstratified holdout partition. The software reserves approximately 30% of the observations for the test data set and uses the rest of the observations for the training data set.
rng("default") % For reproducibility of the partition c = cvpartition(size(tbl,1),"Holdout",0.30); trainingIndices = training(c); testIndices = test(c); tblTrain = tbl(trainingIndices,:); tblTest = tbl(testIndices,:);
Train a regression neural network model using the training set. Specify the Systolic column of tblTrain as the response variable. Specify to standardize the numeric predictors, and set the iteration limit to 50. By default, the neural network model has one fully connected layer with 10 outputs, excluding the final fully connected layer.
Mdl = fitrnet(tblTrain,"Systolic", ... "Standardize",true,"IterationLimit",50);
Predict the systolic blood pressure levels for patients in the test set.
predictedY = predict(Mdl,tblTest);
Visualize the results by using a scatter plot with a reference line. Plot the predicted values along the vertical axis and the true response values along the horizontal axis. Points on the reference line indicate correct predictions.
plot(tblTest.Systolic,predictedY,".") hold on plot(tblTest.Systolic,tblTest.Systolic) hold off xlabel("True Systolic Blood Pressure Levels") ylabel("Predicted Systolic Blood Pressure Levels")
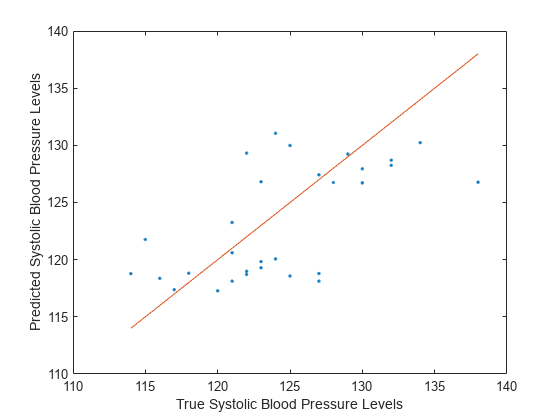
Because many of the points are far from the reference line, the default neural network model with a fully connected layer of size 10 does not seem to be a great predictor of systolic blood pressure levels.
Perform feature selection by comparing test set losses and predictions. Compare the test set metrics for a regression neural network model trained using all the predictors to the test set metrics for a model trained using only a subset of the predictors.
Load the sample file fisheriris.csv, which contains iris data including sepal length, sepal width, petal length, petal width, and species type. Read the file into a table.
fishertable = readtable('fisheriris.csv');Separate the data into a training set trainTbl and a test set testTbl by using a nonstratified holdout partition. The software reserves approximately 30% of the observations for the test data set and uses the rest of the observations for the training data set.
rng("default") c = cvpartition(size(fishertable,1),"Holdout",0.3); trainTbl = fishertable(training(c),:); testTbl = fishertable(test(c),:);
Train one regression neural network model using all the predictors in the training set, and train another model using all the predictors except PetalWidth. For both models, specify PetalLength as the response variable, and standardize the predictors.
allMdl = fitrnet(trainTbl,"PetalLength","Standardize",true); subsetMdl = fitrnet(trainTbl,"PetalLength ~ SepalLength + SepalWidth + Species", ... "Standardize",true);
Compare the test set mean squared error (MSE) of the two models. Smaller MSE values indicate better performance.
allMSE = loss(allMdl,testTbl)
allMSE = 0.0837
subsetMSE = loss(subsetMdl,testTbl)
subsetMSE = 0.0878
For each model, compare the test set predicted petal lengths to the true petal lengths. Plot the predicted petal lengths along the vertical axis and the true petal lengths along the horizontal axis. Points on the reference line indicate correct predictions.
tiledlayout(2,1) % Top axes ax1 = nexttile; allPredictedY = predict(allMdl,testTbl); plot(ax1,testTbl.PetalLength,allPredictedY,".") hold on plot(ax1,testTbl.PetalLength,testTbl.PetalLength) hold off xlabel(ax1,"True Petal Length") ylabel(ax1,"Predicted Petal Length") title(ax1,"All Predictors") % Bottom axes ax2 = nexttile; subsetPredictedY = predict(subsetMdl,testTbl); plot(ax2,testTbl.PetalLength,subsetPredictedY,".") hold on plot(ax2,testTbl.PetalLength,testTbl.PetalLength) hold off xlabel(ax2,"True Petal Length") ylabel(ax2,"Predicted Petal Length") title(ax2,"Subset of Predictors")

Because both models seems to perform well, with predictions scattered near the reference line, consider using the model trained using all predictors except PetalWidth.
See how the layers of a regression neural network model work together to predict the response value for a single observation.
Load the sample file fisheriris.csv, which contains iris data including sepal length, sepal width, petal length, petal width, and species type. Read the file into a table, and display the first few rows of the table.
fishertable = readtable('fisheriris.csv');
head(fishertable) SepalLength SepalWidth PetalLength PetalWidth Species
___________ __________ ___________ __________ __________
5.1 3.5 1.4 0.2 {'setosa'}
4.9 3 1.4 0.2 {'setosa'}
4.7 3.2 1.3 0.2 {'setosa'}
4.6 3.1 1.5 0.2 {'setosa'}
5 3.6 1.4 0.2 {'setosa'}
5.4 3.9 1.7 0.4 {'setosa'}
4.6 3.4 1.4 0.3 {'setosa'}
5 3.4 1.5 0.2 {'setosa'}
Train a regression neural network model using the data set. Specify the PetalLength variable as the response and use the other numeric variables as predictors.
Mdl = fitrnet(fishertable,"PetalLength ~ SepalLength + SepalWidth + PetalWidth");Select the fifteenth observation from the data set. See how the layers of the neural network take the observation and return a predicted response value newPointResponse.
newPoint = Mdl.X{15,:}newPoint = 1×3
5.8000 4.0000 0.2000
firstFCStep = (Mdl.LayerWeights{1})*newPoint' + Mdl.LayerBiases{1};
reluStep = max(firstFCStep,0);
finalFCStep = (Mdl.LayerWeights{end})*reluStep + Mdl.LayerBiases{end};
newPointResponse = finalFCStepnewPointResponse = 1.6716
Check that the prediction matches the one returned by the predict object function.
predictedY = predict(Mdl,newPoint)
predictedY = 1.6716
isequal(newPointResponse,predictedY)
ans = logical
1
The two results match.
Since R2024b
Create a regression neural network with more than one response variable.
Load the carbig data set, which contains measurements of cars made in the 1970s and early 1980s. Create a table containing the predictor variables Displacement, Horsepower, and so on, as well as the response variables Acceleration and MPG. Display the first eight rows of the table.
load carbig cars = table(Displacement,Horsepower,Model_Year, ... Origin,Weight,Acceleration,MPG); head(cars)
Displacement Horsepower Model_Year Origin Weight Acceleration MPG
____________ __________ __________ _______ ______ ____________ ___
307 130 70 USA 3504 12 18
350 165 70 USA 3693 11.5 15
318 150 70 USA 3436 11 18
304 150 70 USA 3433 12 16
302 140 70 USA 3449 10.5 17
429 198 70 USA 4341 10 15
454 220 70 USA 4354 9 14
440 215 70 USA 4312 8.5 14
Remove rows of cars where the table has missing values.
cars = rmmissing(cars);
Categorize the cars based on whether they were made in the USA.
cars.Origin = categorical(cellstr(cars.Origin)); cars.Origin = mergecats(cars.Origin,["France","Japan",... "Germany","Sweden","Italy","England"],"NotUSA");
Partition the data into training and test sets. Use approximately 85% of the observations to train a neural network model, and 15% of the observations to test the performance of the trained model on new data. Use cvpartition to partition the data.
rng("default") % For reproducibility c = cvpartition(height(cars),"Holdout",0.15); carsTrain = cars(training(c),:); carsTest = cars(test(c),:);
Train a multiresponse neural network regression model by passing the carsTrain training data to the fitrnet function. For better results, specify to standardize the predictor data.
Mdl = fitrnet(carsTrain,["Acceleration","MPG"], ... Standardize=true)
Mdl =
RegressionNeuralNetwork
PredictorNames: {'Displacement' 'Horsepower' 'Model_Year' 'Origin' 'Weight'}
ResponseName: {'Acceleration' 'MPG'}
CategoricalPredictors: 4
ResponseTransform: 'none'
NumObservations: 334
LayerSizes: 10
Activations: 'relu'
OutputLayerActivation: 'none'
Solver: 'LBFGS'
ConvergenceInfo: [1×1 struct]
TrainingHistory: [1000×7 table]
Properties, Methods
Mdl is a trained RegressionNeuralNetwork model. You can use dot notation to access the properties of Mdl. For example, you can specify Mdl.ConvergenceInfo to get more information about the model convergence.
Evaluate the performance of the regression model on the test set by computing the test mean squared error (MSE). Smaller MSE values indicate better performance. Return the loss for each response variable separately by setting the OutputType name-value argument to "per-response".
testMSE = loss(Mdl,carsTest,["Acceleration","MPG"], ... OutputType="per-response")
testMSE = 1×2
1.5341 4.8245
Predict the response values for the observations in the test set. Return the predicted response values as a table.
predictedY = predict(Mdl,carsTest,OutputType="table")predictedY=58×2 table
Acceleration MPG
____________ ______
9.3612 13.567
15.655 21.406
17.921 17.851
11.139 13.433
12.696 10.32
16.498 17.977
16.227 22.016
12.165 12.926
12.691 12.072
12.424 14.481
16.974 22.152
15.504 24.955
11.068 13.874
11.978 12.664
14.926 10.134
15.638 24.839
⋮
Input Arguments
Trained regression neural network, specified as a RegressionNeuralNetwork model object or CompactRegressionNeuralNetwork model object returned by fitrnet or
compact,
respectively.
Predictor data used to generate responses, specified as a numeric matrix or table.
By default, each row of X corresponds to one observation, and
each column corresponds to one variable.
For a numeric matrix:
The variables in the columns of
Xmust have the same order as the predictor variables that trainedMdl.If you train
Mdlusing a table (for example,Tbl) andTblcontains only numeric predictor variables, thenXcan be a numeric matrix. To treat numeric predictors inTblas categorical during training, identify categorical predictors by using theCategoricalPredictorsname-value argument offitrnet. IfTblcontains heterogeneous predictor variables (for example, numeric and categorical data types) andXis a numeric matrix, thenpredictthrows an error.
For a table:
predictdoes not support multicolumn variables or cell arrays other than cell arrays of character vectors.If you train
Mdlusing a table (for example,Tbl), then all predictor variables inXmust have the same variable names and data types as the variables that trainedMdl(stored inMdl.PredictorNames). However, the column order ofXdoes not need to correspond to the column order ofTbl. Also,TblandXcan contain additional variables (response variables, observation weights, and so on), butpredictignores them.If you train
Mdlusing a numeric matrix, then the predictor names inMdl.PredictorNamesmust be the same as the corresponding predictor variable names inX. To specify predictor names during training, use thePredictorNamesname-value argument offitrnet. All predictor variables inXmust be numeric vectors.Xcan contain additional variables (response variables, observation weights, and so on), butpredictignores them.
If you set Standardize=true in fitrnet when
training Mdl, then the software standardizes the numeric columns of
the predictor data using the corresponding means (Mdl.Mu) and
standard deviations (Mdl.Sigma).
Note
If you orient your predictor matrix so that observations correspond to columns and
specify ObservationsIn="columns", then you might experience a
significant reduction in computation time. You cannot specify
ObservationsIn="columns" for predictor data in a table or for
multiresponse regression.
Data Types: single | double | table
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Example: predict(Mdl,X,ObservationsIn="columns") indicates that
columns in the predictor data correspond to observations.
Predictor data observation dimension, specified as "rows" or
"columns".
Note
If you orient your predictor matrix so that observations correspond to columns
and specify ObservationsIn="columns", then you might experience a
significant reduction in computation time. You cannot specify
ObservationsIn="columns" for predictor data in a table or for
multiresponse regression.
Data Types: char | string
Since R2023b
Predicted response value to use for observations with missing predictor values, specified as "median", "mean", or a numeric scalar.
| Value | Description |
|---|---|
"median" | predict uses the median of the observed response values in the training data as the predicted response value for observations with missing predictor values. |
"mean" | predict uses the mean of the observed response values in the training data as the predicted response value for observations with missing predictor values. |
| Numeric scalar | predict uses this value as the predicted response value for observations with missing predictor values. |
Example: PredictionForMissingValue="mean"
Example: PredictionForMissingValue=NaN
Data Types: single | double | char | string
Output Arguments
Predicted responses, returned as a numeric vector, matrix, or table.
If
yfitis a numeric vector, then entry i inyfitcorresponds to observation i inX.If
yfitis a numeric matrix or table, then row i inyfitcorresponds to observation i inX.
Alternative Functionality
Simulink Block
To integrate the prediction of a neural network regression model into Simulink®, you can use the RegressionNeuralNetwork
Predict block in the Statistics and Machine Learning Toolbox™ library or a MATLAB® Function block with the predict function. For examples,
see Predict Responses Using RegressionNeuralNetwork Predict Block and Predict Class Labels Using MATLAB Function Block.
When deciding which approach to use, consider the following:
If you use the Statistics and Machine Learning Toolbox library block, you can use the Fixed-Point Tool (Fixed-Point Designer) to convert a floating-point model to fixed point.
Support for variable-size arrays must be enabled for a MATLAB Function block with the
predictfunction.If you use a MATLAB Function block, you can use MATLAB functions for preprocessing or post-processing before or after predictions in the same MATLAB Function block.
Extended Capabilities
Usage notes and limitations:
Use
saveLearnerForCoder,loadLearnerForCoder, andcodegen(MATLAB Coder) to generate code for thepredictfunction. Save a trained model by usingsaveLearnerForCoder. Define an entry-point function that loads the saved model by usingloadLearnerForCoderand calls thepredictfunction. Then usecodegento generate code for the entry-point function. Code generation is not supported for multiresponse regression.To generate single-precision C/C++ code for
predict, specifyDataType="single"when you call theloadLearnerForCoderfunction.This table contains notes about the arguments of
predict. Arguments not included in this table are fully supported.Argument Notes and Limitations MdlFor the usage notes and limitations of the model object, see Code Generation of the
CompactRegressionNeuralNetworkobject.XXmust be a single-precision or double-precision matrix or a table containing numeric variables, categorical variables, or both.The number of rows, or observations, in
Xcan be a variable size, but the number of columns inXmust be fixed.If you want to specify
Xas a table, then your model must be trained using a table, and your entry-point function for prediction must do the following:Accept data as arrays.
Create a table from the data input arguments and specify the variable names in the table.
Pass the table to
predict.
For an example of this table workflow, see Generate Code to Classify Data in Table. For more information on using tables in code generation, see Code Generation for Tables (MATLAB Coder) and Table Limitations for Code Generation (MATLAB Coder).
Name-value arguments Names in name-value arguments must be compile-time constants.
The
ObservationsInvalue must be a compile-time constant. For example, to useObservationsIn="columns"in the generated code, include{coder.Constant("ObservationsIn"),coder.Constant("columns")}in the-argsvalue ofcodegen(MATLAB Coder).The
OutputTypename-value argument is not supported for code generation.If the value of
PredictionForMissingValueis nonnumeric, then it must be a compile-time constant.
For more information, see Introduction to Code Generation.
This function fully supports GPU arrays. For more information, see Run MATLAB Functions on a GPU (Parallel Computing Toolbox).
Version History
Introduced in R2021aYou can create a neural network regression model with multiple response variables by
using the fitrnet function.
Regardless of the number of response variables, the function returns a
RegressionNeuralNetwork object. You can use the
predict object function to predict the responses for new data.
In the call to predict, you can specify whether to return the
predicted response values as a matrix or table by using the OutputType
name-value argument.
predict fully supports GPU arrays.
Starting in R2023b, when you predict or compute the loss, some regression models allow you to specify the predicted response value for observations with missing predictor values. Specify the PredictionForMissingValue name-value argument to use a numeric scalar, the training set median, or the training set mean as the predicted value. When computing the loss, you can also specify to omit observations with missing predictor values.
This table lists the object functions that support the
PredictionForMissingValue name-value argument. By default, the
functions use the training set median as the predicted response value for observations with
missing predictor values.
| Model Type | Model Objects | Object Functions |
|---|---|---|
| Gaussian process regression (GPR) model | RegressionGP, CompactRegressionGP | loss, predict, resubLoss, resubPredict |
RegressionPartitionedGP | kfoldLoss, kfoldPredict | |
| Gaussian kernel regression model | RegressionKernel | loss, predict |
RegressionPartitionedKernel | kfoldLoss, kfoldPredict | |
| Linear regression model | RegressionLinear | loss, predict |
RegressionPartitionedLinear | kfoldLoss, kfoldPredict | |
| Neural network regression model | RegressionNeuralNetwork, CompactRegressionNeuralNetwork | loss, predict, resubLoss, resubPredict |
RegressionPartitionedNeuralNetwork | kfoldLoss, kfoldPredict | |
| Support vector machine (SVM) regression model | RegressionSVM, CompactRegressionSVM | loss, predict, resubLoss, resubPredict |
RegressionPartitionedSVM | kfoldLoss, kfoldPredict |
In previous releases, the regression model loss and predict functions listed above used NaN predicted response values for observations with missing predictor values. The software omitted observations with missing predictor values from the resubstitution ("resub") and cross-validation ("kfold") computations for prediction and loss.
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
웹사이트 선택
번역된 콘텐츠를 보고 지역별 이벤트와 혜택을 살펴보려면 웹사이트를 선택하십시오. 현재 계신 지역에 따라 다음 웹사이트를 권장합니다:
또한 다음 목록에서 웹사이트를 선택하실 수도 있습니다.
사이트 성능 최적화 방법
최고의 사이트 성능을 위해 중국 사이트(중국어 또는 영어)를 선택하십시오. 현재 계신 지역에서는 다른 국가의 MathWorks 사이트 방문이 최적화되지 않았습니다.
미주
- América Latina (Español)
- Canada (English)
- United States (English)
유럽
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)