segmentObjects
Syntax
Description
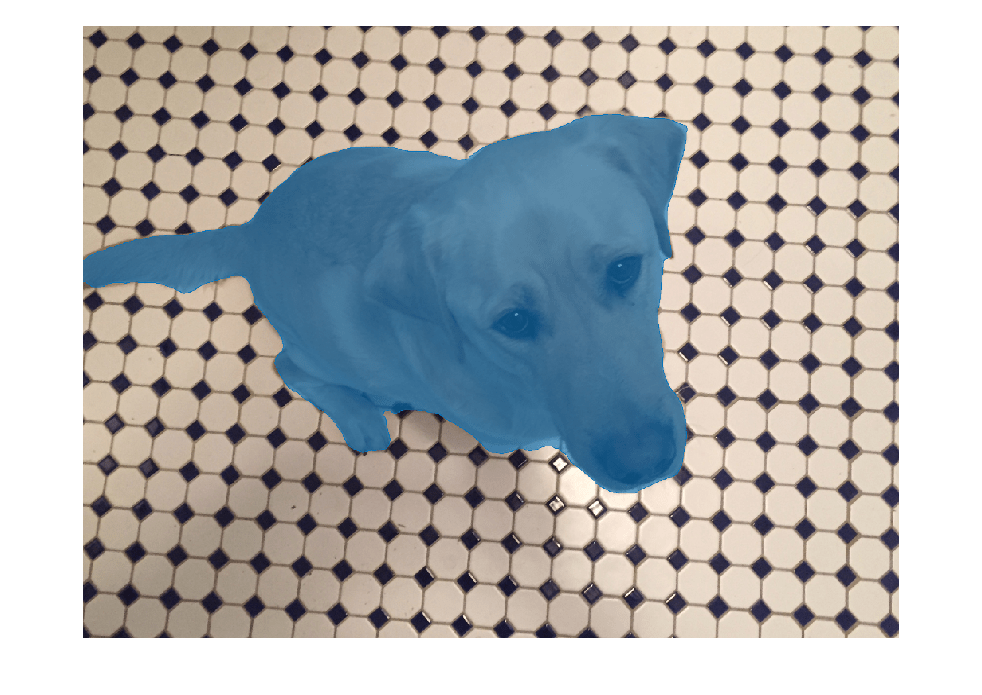
masks = segmentObjects(detector,I)I using
SOLOv2 instance segmentation, and returns the predicted object masks for the input image
or images.
Note
This functionality requires Deep Learning Toolbox™ and the Computer Vision Toolbox™ Model for SOLOv2 Instance Segmentation. You can install the Computer Vision Toolbox Model for SOLOv2 Instance Segmentation from Add-On Explorer. For more information about installing add-ons, see Get and Manage Add-Ons.
[___] = segmentObjects(___,
specifies options using additional name-value arguments in addition to any combination of
arguments from previous syntaxes.. For example, Name=Value)Threshold=0.9 specifies
the confidence threshold as 0.9.