loss
Class: ClassificationLinear
Classification loss for linear classification models
Description
L = loss(Mdl,Tbl,ResponseVarName)Tbl and the true class labels in
Tbl.ResponseVarName.
L = loss(___,Name,Value)
Note
If the predictor data in X or Tbl contains
any missing values and LossFun is not set to
"classifcost", "classiferror", or
"mincost", the loss function can
return NaN. For more details, see loss can return NaN for predictor data with missing values.
Input Arguments
Binary, linear classification model, specified as a ClassificationLinear model object.
You can create a ClassificationLinear model object
using fitclinear.
Predictor data, specified as an n-by-p full or sparse matrix. This orientation of X indicates that rows correspond to individual observations, and columns correspond to individual predictor variables.
Note
If you orient your predictor matrix so that observations correspond to columns and specify 'ObservationsIn','columns', then you might experience a significant reduction in computation time.
The length of Y and the number of observations
in X must be equal.
Data Types: single | double
Class labels, specified as a categorical, character, or string array; logical or numeric vector; or cell array of character vectors.
The data type of
Ymust be the same as the data type ofMdl.ClassNames. (The software treats string arrays as cell arrays of character vectors.)The distinct classes in
Ymust be a subset ofMdl.ClassNames.If
Yis a character array, then each element must correspond to one row of the array.The length of
Ymust be equal to the number of observations inXorTbl.
Data Types: categorical | char | string | logical | single | double | cell
Sample data used to train the model, specified as a table. Each row of
Tbl corresponds to one observation, and each column corresponds
to one predictor variable. Optionally, Tbl can contain additional
columns for the response variable and observation weights. Tbl must
contain all the predictors used to train Mdl. Multicolumn variables
and cell arrays other than cell arrays of character vectors are not allowed.
If Tbl contains the response variable used to train Mdl, then you do not need to specify ResponseVarName or Y.
If you train Mdl using sample data contained in a table, then the input
data for loss must also be in a table.
Response variable name, specified as the name of a variable in Tbl. If Tbl contains the response variable used to train Mdl, then you do not need to specify ResponseVarName.
If you specify ResponseVarName, then you must specify it as a character
vector or string scalar. For example, if the response variable is stored as
Tbl.Y, then specify ResponseVarName as
'Y'. Otherwise, the software treats all columns of
Tbl, including Tbl.Y, as predictors.
The response variable must be a categorical, character, or string array; a logical or numeric vector; or a cell array of character vectors. If the response variable is a character array, then each element must correspond to one row of the array.
Data Types: char | string
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Before R2021a, use commas to separate each name and value, and enclose
Name in quotes.
Loss function, specified as the comma-separated pair consisting
of 'LossFun' and a built-in loss function name
or function handle.
The following table lists the available loss functions. Specify one using its corresponding character vector or string scalar.
Value Description "binodeviance"Binomial deviance "classifcost"Observed misclassification cost "classiferror"Misclassified rate in decimal "exponential"Exponential loss "hinge"Hinge loss "logit"Logistic loss "mincost"Minimal expected misclassification cost (for classification scores that are posterior probabilities) "quadratic"Quadratic loss 'mincost'is appropriate for classification scores that are posterior probabilities. For linear classification models, logistic regression learners return posterior probabilities as classification scores by default, but SVM learners do not (seepredict).To specify a custom loss function, use function handle notation. The function must have this form:
lossvalue =lossfun(C,S,W,Cost)The output argument
lossvalueis a scalar.You specify the function name (
lossfun).Cis ann-by-Klogical matrix with rows indicating the class to which the corresponding observation belongs.nis the number of observations inTblorX, andKis the number of distinct classes (numel(Mdl.ClassNames)). The column order corresponds to the class order inMdl.ClassNames. CreateCby settingC(p,q) = 1, if observationpis in classq, for each row. Set all other elements of rowpto0.Sis ann-by-Knumeric matrix of classification scores. The column order corresponds to the class order inMdl.ClassNames.Sis a matrix of classification scores, similar to the output ofpredict.Wis ann-by-1 numeric vector of observation weights.Costis aK-by-Knumeric matrix of misclassification costs. For example,Cost = ones(K) – eye(K)specifies a cost of0for correct classification and1for misclassification.
Example: 'LossFun',@lossfun
Data Types: char | string | function_handle
Predictor data observation dimension, specified as 'rows' or
'columns'.
Note
If you orient your predictor matrix so that observations correspond to columns and
specify 'ObservationsIn','columns', then you might experience a
significant reduction in computation time. You cannot specify
'ObservationsIn','columns' for predictor data in a
table.
Data Types: char | string
Observation weights, specified as the comma-separated pair consisting
of 'Weights' and a numeric vector or the name of a
variable in Tbl.
If you specify
Weightsas a numeric vector, then the size ofWeightsmust be equal to the number of observations inXorTbl.If you specify
Weightsas the name of a variable inTbl, then the name must be a character vector or string scalar. For example, if the weights are stored asTbl.W, then specifyWeightsas'W'. Otherwise, the software treats all columns ofTbl, includingTbl.W, as predictors.
If you supply weights, then for each regularization strength, loss
computes the weighted classification loss and
normalizes weights to sum up to the value of the prior probability in
the respective class.
Data Types: double | single
Output Arguments
Examples
Load the NLP data set.
load nlpdataX is a sparse matrix of predictor data, and Y is a categorical vector of class labels. There are more than two classes in the data.
The models should identify whether the word counts in a web page are from the Statistics and Machine Learning Toolbox™ documentation. So, identify the labels that correspond to the Statistics and Machine Learning Toolbox™ documentation web pages.
Ystats = Y == 'stats';Train a binary, linear classification model that can identify whether the word counts in a documentation web page are from the Statistics and Machine Learning Toolbox™ documentation. Specify to hold out 30% of the observations. Optimize the objective function using SpaRSA.
rng(1); % For reproducibility CVMdl = fitclinear(X,Ystats,'Solver','sparsa','Holdout',0.30); CMdl = CVMdl.Trained{1};
CVMdl is a ClassificationPartitionedLinear model. It contains the property Trained, which is a 1-by-1 cell array holding a ClassificationLinear model that the software trained using the training set.
Extract the training and test data from the partition definition.
trainIdx = training(CVMdl.Partition); testIdx = test(CVMdl.Partition);
Estimate the training- and test-sample classification error.
ceTrain = loss(CMdl,X(trainIdx,:),Ystats(trainIdx))
ceTrain = 1.3572e-04
ceTest = loss(CMdl,X(testIdx,:),Ystats(testIdx))
ceTest = 5.2804e-04
Because there is one regularization strength in CMdl, ceTrain and ceTest are numeric scalars.
Load the NLP data set. Preprocess the data as in Estimate Test-Sample Classification Loss, and transpose the predictor data.
load nlpdata Ystats = Y == 'stats'; X = X';
Train a binary, linear classification model. Specify to hold out 30% of the observations. Optimize the objective function using SpaRSA. Specify that the predictor observations correspond to columns.
rng(1); % For reproducibility CVMdl = fitclinear(X,Ystats,'Solver','sparsa','Holdout',0.30,... 'ObservationsIn','columns'); CMdl = CVMdl.Trained{1};
CVMdl is a ClassificationPartitionedLinear model. It contains the property Trained, which is a 1-by-1 cell array holding a ClassificationLinear model that the software trained using the training set.
Extract the training and test data from the partition definition.
trainIdx = training(CVMdl.Partition); testIdx = test(CVMdl.Partition);
Create an anonymous function that measures linear loss, that is,
is the weight for observation j, is response j (-1 for the negative class, and 1 otherwise), and is the raw classification score of observation j. Custom loss functions must be written in a particular form. For rules on writing a custom loss function, see the LossFun name-value pair argument.
linearloss = @(C,S,W,Cost)sum(-W.*sum(S.*C,2))/sum(W);
Estimate the training- and test-sample classification loss using the linear loss function.
ceTrain = loss(CMdl,X(:,trainIdx),Ystats(trainIdx),'LossFun',linearloss,... 'ObservationsIn','columns')
ceTrain = -7.8330
ceTest = loss(CMdl,X(:,testIdx),Ystats(testIdx),'LossFun',linearloss,... 'ObservationsIn','columns')
ceTest = -7.7383
To determine a good lasso-penalty strength for a linear classification model that uses a logistic regression learner, compare test-sample classification error rates.
Load the NLP data set. Preprocess the data as in Specify Custom Classification Loss.
load nlpdata Ystats = Y == 'stats'; X = X'; rng(10); % For reproducibility Partition = cvpartition(Ystats,'Holdout',0.30); testIdx = test(Partition); XTest = X(:,testIdx); YTest = Ystats(testIdx);
Create a set of 11 logarithmically-spaced regularization strengths from through .
Lambda = logspace(-6,-0.5,11);
Train binary, linear classification models that use each of the regularization strengths. Optimize the objective function using SpaRSA. Lower the tolerance on the gradient of the objective function to 1e-8.
CVMdl = fitclinear(X,Ystats,'ObservationsIn','columns',... 'CVPartition',Partition,'Learner','logistic','Solver','sparsa',... 'Regularization','lasso','Lambda',Lambda,'GradientTolerance',1e-8)
CVMdl =
ClassificationPartitionedLinear
CrossValidatedModel: 'Linear'
ResponseName: 'Y'
NumObservations: 31572
KFold: 1
Partition: [1×1 cvpartition]
ClassNames: [0 1]
ScoreTransform: 'none'
Properties, Methods
Extract the trained linear classification model.
Mdl = CVMdl.Trained{1}Mdl =
ClassificationLinear
ResponseName: 'Y'
ClassNames: [0 1]
ScoreTransform: 'logit'
Beta: [34023×11 double]
Bias: [-12.1623 -12.1623 -12.1623 -12.1623 -12.1623 -6.2503 -5.0651 -4.2165 -3.3990 -3.2452 -2.9783]
Lambda: [1.0000e-06 3.5481e-06 1.2589e-05 4.4668e-05 1.5849e-04 5.6234e-04 0.0020 0.0071 0.0251 0.0891 0.3162]
Learner: 'logistic'
Properties, Methods
Mdl is a ClassificationLinear model object. Because Lambda is a sequence of regularization strengths, you can think of Mdl as 11 models, one for each regularization strength in Lambda.
Estimate the test-sample classification error.
ce = loss(Mdl,X(:,testIdx),Ystats(testIdx),'ObservationsIn','columns');
Because there are 11 regularization strengths, ce is a 1-by-11 vector of classification error rates.
Higher values of Lambda lead to predictor variable sparsity, which is a good quality of a classifier. For each regularization strength, train a linear classification model using the entire data set and the same options as when you cross-validated the models. Determine the number of nonzero coefficients per model.
Mdl = fitclinear(X,Ystats,'ObservationsIn','columns',... 'Learner','logistic','Solver','sparsa','Regularization','lasso',... 'Lambda',Lambda,'GradientTolerance',1e-8); numNZCoeff = sum(Mdl.Beta~=0);
In the same figure, plot the test-sample error rates and frequency of nonzero coefficients for each regularization strength. Plot all variables on the log scale.
figure; [h,hL1,hL2] = plotyy(log10(Lambda),log10(ce),... log10(Lambda),log10(numNZCoeff + 1)); hL1.Marker = 'o'; hL2.Marker = 'o'; ylabel(h(1),'log_{10} classification error') ylabel(h(2),'log_{10} nonzero-coefficient frequency') xlabel('log_{10} Lambda') title('Test-Sample Statistics') hold off
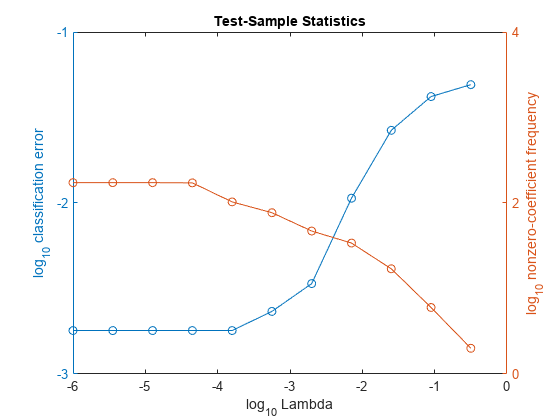
Choose the index of the regularization strength that balances predictor variable sparsity and low classification error. In this case, a value between to should suffice.
idxFinal = 7;
Select the model from Mdl with the chosen regularization strength.
MdlFinal = selectModels(Mdl,idxFinal);
MdlFinal is a ClassificationLinear model containing one regularization strength. To estimate labels for new observations, pass MdlFinal and the new data to predict.
More About
Classification loss functions measure the predictive inaccuracy of classification models. When you compare the same type of loss among many models, a lower loss indicates a better predictive model.
Consider the following scenario.
L is the weighted average classification loss.
n is the sample size.
yj is the observed class label. The software codes it as –1 or 1, indicating the negative or positive class (or the first or second class in the
ClassNamesproperty), respectively.f(Xj) is the positive-class classification score for observation (row) j of the predictor data X.
mj = yjf(Xj) is the classification score for classifying observation j into the class corresponding to yj. Positive values of mj indicate correct classification and do not contribute much to the average loss. Negative values of mj indicate incorrect classification and contribute significantly to the average loss.
The weight for observation j is wj. The software normalizes the observation weights so that they sum to the corresponding prior class probability stored in the
Priorproperty. Therefore,
Given this scenario, the following table describes the supported loss functions that you can specify by using the LossFun name-value argument.
| Loss Function | Value of LossFun | Equation |
|---|---|---|
| Binomial deviance | "binodeviance" | |
| Observed misclassification cost | "classifcost" | where is the class label corresponding to the class with the maximal score, and is the user-specified cost of classifying an observation into class when its true class is yj. |
| Misclassified rate in decimal | "classiferror" | where I{·} is the indicator function. |
| Cross-entropy loss | "crossentropy" |
The weighted cross-entropy loss is where the weights are normalized to sum to n instead of 1. |
| Exponential loss | "exponential" | |
| Hinge loss | "hinge" | |
| Logistic loss | "logit" | |
| Minimal expected misclassification cost | "mincost" |
The software computes the weighted minimal expected classification cost using this procedure for observations j = 1,...,n.
The weighted average of the minimal expected misclassification cost loss is |
| Quadratic loss | "quadratic" |
If you use the default cost matrix (whose element value is 0 for correct classification
and 1 for incorrect classification), then the loss values for
"classifcost", "classiferror", and
"mincost" are identical. For a model with a nondefault cost matrix,
the "classifcost" loss is equivalent to the "mincost"
loss most of the time. These losses can be different if prediction into the class with
maximal posterior probability is different from prediction into the class with minimal
expected cost. Note that "mincost" is appropriate only if classification
scores are posterior probabilities.
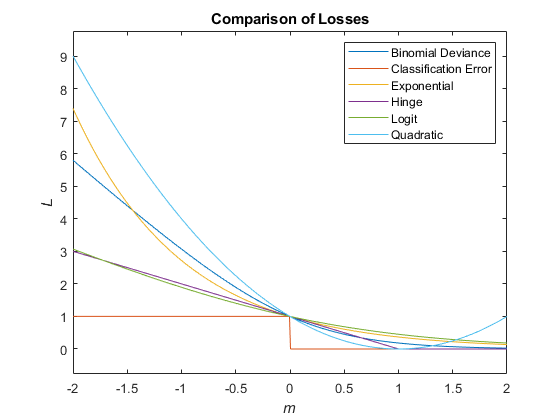
This figure compares the loss functions (except "classifcost",
"crossentropy", and "mincost") over the score
m for one observation. Some functions are normalized to pass through
the point (0,1).
Algorithms
By default, observation weights are prior class probabilities.
If you supply weights using Weights, then the
software normalizes them to sum to the prior probabilities in the
respective classes. The software uses the renormalized weights to
estimate the weighted classification loss.
Extended Capabilities
The
loss function supports tall arrays with the following usage
notes and limitations:
lossdoes not support talltabledata.
For more information, see Tall Arrays.
This function fully supports GPU arrays. For more information, see Run MATLAB Functions on a GPU (Parallel Computing Toolbox).
Version History
Introduced in R2016aloss fully supports GPU arrays.
If you specify a nondefault cost matrix when you train the input model object, the loss function returns a different value compared to previous releases.
The loss function uses the prior
probabilities stored in the Prior property to normalize the observation
weights of the input data. Also, the function uses the cost matrix stored in the
Cost property if you specify the LossFun name-value
argument as "classifcost" or "mincost". The way the
function uses the Prior and Cost property values has not
changed. However, the property values stored in the input model object have changed for a model
with a nondefault cost matrix, so the function might return a different value.
For details about the property value change, see Cost property stores the user-specified cost matrix.
If you want the software to handle the cost matrix, prior
probabilities, and observation weights in the same way as in previous releases, adjust the prior
probabilities and observation weights for the nondefault cost matrix, as described in Adjust Prior Probabilities and Observation Weights for Misclassification Cost Matrix. Then, when you train a
classification model, specify the adjusted prior probabilities and observation weights by using
the Prior and Weights name-value arguments, respectively,
and use the default cost matrix.
The loss function no longer omits an observation with a
NaN score when computing the weighted average classification loss. Therefore,
loss can now return NaN when the predictor data
X or the predictor variables in Tbl
contain any missing values, and the name-value argument LossFun is
not specified as "classifcost", "classiferror", or
"mincost". In most cases, if the test set observations do not
contain missing predictors, the loss function does not
return NaN.
This change improves the automatic selection of a classification model when you use
fitcauto.
Before this change, the software might select a model (expected to best classify new
data) with few non-NaN predictors.
If loss in your code returns NaN, you can update your code
to avoid this result by doing one of the following:
Remove or replace the missing values by using
rmmissingorfillmissing, respectively.Specify the name-value argument
LossFunas"classifcost","classiferror", or"mincost".
The following table shows the classification models for which the
loss object function might return NaN. For more details,
see the Compatibility Considerations for each loss
function.
| Model Type | Full or Compact Model Object | loss Object
Function |
|---|---|---|
| Discriminant analysis classification model | ClassificationDiscriminant, CompactClassificationDiscriminant | loss |
| Ensemble of learners for classification | ClassificationEnsemble, CompactClassificationEnsemble | loss |
| Gaussian kernel classification model | ClassificationKernel | loss |
| k-nearest neighbor classification model | ClassificationKNN | loss |
| Linear classification model | ClassificationLinear | loss |
| Neural network classification model | ClassificationNeuralNetwork, CompactClassificationNeuralNetwork | loss |
| Support vector machine (SVM) classification model | loss |
See Also
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
웹사이트 선택
번역된 콘텐츠를 보고 지역별 이벤트와 혜택을 살펴보려면 웹사이트를 선택하십시오. 현재 계신 지역에 따라 다음 웹사이트를 권장합니다:
또한 다음 목록에서 웹사이트를 선택하실 수도 있습니다.
사이트 성능 최적화 방법
최고의 사이트 성능을 위해 중국 사이트(중국어 또는 영어)를 선택하십시오. 현재 계신 지역에서는 다른 국가의 MathWorks 사이트 방문이 최적화되지 않았습니다.
미주
- América Latina (Español)
- Canada (English)
- United States (English)
유럽
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)