Calculate Sensitivities Using sbiosimulate
This example performs local sensitivity analysis on a Model of the Yeast Heterotrimeric G Protein Cycle to find parameters that influence the amount of active G protein. Assume that you are calculating the sensitivity of species Ga with respect to every parameter in the model. Thus, you want to calculate the time-dependent derivatives:
Load G-protein Model
The provided SimBiology project gprotein_norules.sbproj contains a model that represents the wild-type strain (stored in variable m1).
sbioloadproject gprotein_norules.sbproj m1
Set up Sensitivity Analysis Options
The options for sensitivity analysis are in the configset object of the model.
csObj = getconfigset(m1);
Use the sbioselect function, which lets you query by type, to retrieve the Ga species from the model.
Ga = sbioselect(m1,'Name','Ga');
Set the Outputs property of the SensitivityAnalysisOptions object to the Ga species.
csObj.SensitivityAnalysisOptions.Outputs = Ga;
Retrieve all the parameters from the model and store the vector in a variable, pif.
pif = sbioselect(m1,'Type','parameter');
Set the Inputs property of the SensitivityAnalysisOptions object to the pif variable containing the parameters.
csObj.SensitivityAnalysisOptions.Inputs = pif;
Enable sensitivity analysis in the configset object by setting the SensitivityAnalysis option to true.
csObj.SolverOptions.SensitivityAnalysis = true;
Set the Normalization property of the SensitivityAnalysisOptions object to perform 'Full' normalization.
csObj.SensitivityAnalysisOptions.Normalization = 'Full';Calculate Sensitivities
Simulate the model and return the data to a SimData object:
simDataObj = sbiosimulate(m1);
Extract and Plot Sensitivity Data
You can extract sensitivity results using the getsensmatrix method of a SimData object. In this example, R is the sensitivity of the species Ga with respect to eight parameters. This example shows how to compare the variation of sensitivity of Ga with respect to various parameters, and find the parameters that affect Ga the most.
[T, R, snames, ifacs] = getsensmatrix(simDataObj);
Because R is a 3-D array with dimensions corresponding to times, output factors, and input factors, reshape R into columns of input factors to facilitate visualization and plotting:
R2 = squeeze(R);
After extracting the data and reshaping the matrix, plot the data:
figure; plot(T,R2); title('Normalized Sensitivity of Ga With Respect To Various Parameters'); xlabel('Time (seconds)'); ylabel('Normalized Sensitivity of Ga'); leg = legend(ifacs, 'Location', 'NorthEastOutside'); set(leg, 'Interpreter', 'none');
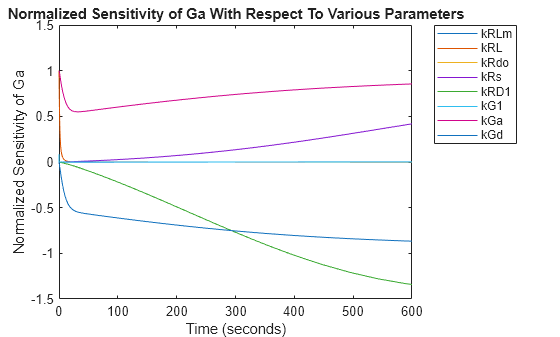
From the plot you can see that Ga is most sensitive to parameters kGd, kRs, kRD1, and kGa. This suggests that the amounts of active G protein in the cell depends on the rate of:
Receptor synthesis
Degradation of the receptor-ligand complex
G protein activation
G protein inactivation
See Also
SimFunctionSensitivity | sbiosimulate | createSimFunction