incrementalOneClassSVM
One-class support vector machine (SVM) model for incremental anomaly detection
Since R2023b
Description
The incrementalOneClassSVM function creates an
incrementalOneClassSVM model object, which represents a one-class SVM model for
incremental anomaly detection.
Unlike other Statistics and Machine Learning Toolbox™ model objects, incrementalOneClassSVM can be called directly. Also,
you can specify learning options, such as the mini-batch size for each learning cycle, the
learning rate, and whether to standardize the predictor data before fitting the model to data.
After you create an incrementalOneClassSVM object, it is prepared for incremental
learning (see Incremental Learning for Anomaly Detection).
incrementalOneClassSVM is best suited for incremental learning. For a traditional
approach to anomaly detection when all the data is provided in advance, see ocsvm.
Note
Incremental learning functions support only numeric input predictor data. You must
prepare an encoded version of categorical data to use incremental learning functions. Use
dummyvar to convert each categorical variable
to a dummy variable. For more details, see Dummy Variables.
Creation
You can create an incrementalOneClassSVM model object in several ways:
Call the function directly — Configure incremental learning options, or specify learner-specific options, by calling
incrementalOneClassSVMdirectly. This approach is best when you do not have data yet or you want to start incremental learning immediately.Convert a traditionally trained model — To initialize a one-class SVM model for incremental learning using the model parameters of a trained
OneClassSVMmodel object, you can convert the traditionally trained model to anincrementalOneClassSVMmodel object by passing it to theincrementalLearnerfunction.Call an incremental learning function —
fitaccepts a configuredincrementalOneClassSVMmodel object and data as input, and returns anincrementalOneClassSVMmodel object updated with information learned from the input model and data.
Description
Mdl = incrementalOneClassSVMMdl for anomaly
detection. Properties of a default model contain placeholders for unknown model
parameters. You must train a default model before you can use it to detect
anomalies.
Mdl = incrementalOneClassSVM(Name=Value)incrementalOneClassSVM(ContaminationFraction=0.1,ScoreWarmupPeriod=1000)
sets the anomaly contamination fraction to 0.1 and the score warm-up
period to 1000.
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Example: Shuffle=true,StandardizeData=true specifies to shuffle the
observations at each iteration, and to standardize the predictor data.
Random number stream for reproducibility of data transformation, specified as a random stream object. For details, see Random Feature Expansion.
Use RandomStream to reproduce the random basis functions used
by incrementalOneClassSVM to transform the data in X to
a high-dimensional space. For details, see Managing the Global Stream Using RandStream and Creating and Controlling a Random Number Stream.
Example: RandomStream=RandStream("mlfg6331_64")
This property is read-only.
Flag for shuffling the observations at each iteration, specified as a value in this table.
| Value | Description |
|---|---|
1 (true) | The software shuffles the observations in an incoming chunk of
data before the fit function fits the model. This
action reduces bias induced by the sampling scheme. |
0 (false) | The software processes the data in the order received. |
This option is valid only when Solver is
"scale-invariant". When Solver is
"sgd" or "asgd", the software always shuffles
the observations in an incoming chunk of data before processing the data.
Example: Shuffle=false
Data Types: logical
Flag to standardize the predictor data, specified as a value in this table.
| Value | Description |
|---|---|
"auto" | incrementalOneClassSVM determines whether the predictor
variables need to be standardized. |
1 (true) | The software standardizes the predictor data. |
0 (false) | The software does not standardize the predictor data. |
Under some conditions, incrementalOneClassSVM can override your
specification. For more details, see Standardize Data.
Example: StandardizeData=true
Data Types: logical | char | string
Properties
You can set most properties by using name-value argument syntax when you call
incrementalOneClassSVM directly. You can set some properties when you call
incrementalLearner to convert a traditionally trained model object. You
cannot set the properties FittedLoss, Mu,
NumTrainingObservations, ScoreThreshold,
Sigma, SolverOptions, and
IsWarm.
One-Class SVM Model Parameters
This property is read-only.
Fraction of anomalies in the training data, specified as a numeric scalar in the
range [0,1].
If the
ContaminationFractionvalue is0(default), thenincrementalOneClassSVMtreats all training observations as normal observations, and sets the score threshold (ScoreThresholdproperty value) to the maximum anomaly score value of the training data.If the
ContaminationFractionvalue is in the range (0,1], thenincrementalOneClassSVMdetermines the threshold value (ScoreThresholdproperty value) so that the function detects the specified fraction of training observations as anomalies.
The default ContaminationFraction value depends on how you
create the model:
If you convert a traditionally trained model to create
Mdl,ContaminationFractionis specified by the corresponding property of the traditionally trained model.If you create
Mdlby callingincrementalOneClassSVMdirectly, you can specifyContaminationFractionby using name-value argument syntax. If you do not specify the value, then the default value is0.
Data Types: single | double
This property is read-only.
Loss function used to fit the linear model, returned as
"hinge". The function has the form .
This property is read-only.
Kernel scale parameter, specified as "auto" or a positive scalar.
incrementalOneClassSVM stores the KernelScale value
as a numeric scalar. The software obtains a random basis for feature expansion by using
the kernel scale parameter. For details, see Random Feature Expansion.
If you specify "auto" when creating the model object, the software
selects an appropriate kernel scale parameter using a heuristic procedure. This
procedure uses subsampling, so estimates might vary from one call to another. Therefore,
to reproduce results, set a random number seed by using rng before training.
The default KernelScale value depends on how you create the model:
If you convert a traditionally trained model to create
Mdl,KernelScaleis specified by the corresponding property of the traditionally trained model.Otherwise, the default value is
1.
Data Types: char | string | single | double
This property is read-only.
Predictor means, specified as a numeric vector.
If Mu is an empty array [] and you specify
StandardizeData=true, the incremental fitting function fit sets
Mu to the predictor variable means estimated during the
estimation period specified by EstimationPeriod.
You cannot specify Mu directly.
Data Types: single | double
This property is read-only.
Number of dimensions of the expanded space, specified as "auto"
or a positive integer. incrementalOneClassSVM stores the
NumExpansionDimensions value as a numeric scalar.
For "auto", the software selects the number of dimensions using
2.^ceil(min(log2(p)+5,15)), where p is the
number of predictors. For details, see Random Feature Expansion.
The default NumExpansionDimensions value depends on how you
create the model:
If you convert a traditionally trained model to create
Mdl,NumExpansionDimensionsis specified by the corresponding property of the traditionally trained model.Otherwise, the default value is
"auto".
Data Types: char | string | single | double
This property is read-only.
Number of predictor variables, specified as a nonnegative numeric scalar.
The default NumPredictors value depends on how you create the model:
If you convert a traditionally trained model to create
Mdl,NumPredictorsis specified by the corresponding property of the traditionally trained model.If you create
Mdlby callingincrementalOneClassSVMdirectly, you can specifyNumPredictorsby using name-value argument syntax. If you do not specify the value, then the default value is0, and incremental fitting functions inferNumPredictorsfrom the predictor data during training.
Data Types: double
This property is read-only.
Number of observations fit to the incremental model Mdl,
specified as a nonnegative numeric scalar. NumTrainingObservations
increases when you pass Mdl and training data to
fit outside of the estimation period.
When fitting the model, the software ignores observations that contain at least one missing value.
If you convert a traditionally trained model to create
Mdl,incrementalOneClassSVMdoes not add the number of observations fit to the traditionally trained model toNumTrainingObservations.
You cannot specify NumTrainingObservations directly.
Data Types: double
This property is read-only.
Predictor variable names, specified as a cell array of character vectors. The
order of the elements in PredictorNames corresponds to the order
in which the predictor names appear in the training data. If the training data is in a
table TBL, the predictor variable names must be a subset of the
variable names in TBL, and the fit and
isanomaly
functions use only the selected variables. The software infers
NumPredictors based on the value of
PredictorNames.
Data Types: cell
This property is read-only.
Predictor standard deviations, specified as a numeric vector.
If Sigma is an empty array [] and you
specify StandardizeData=true, the incremental fitting function
fit sets
Sigma to the predictor variable standard deviations estimated
during the estimation period specified by EstimationPeriod.
You cannot specify Sigma directly.
Data Types: single | double
SGD and ASGD Solver Parameters
This property is read-only.
Mini-batch size for the stochastic solvers, specified as a positive integer. You
cannot specify this parameter when Solver is
"scale-invariant".
At each learning cycle during training, incrementalOneClassSVM uses
BatchSize observations to compute the subgradient. The number of
observations for the last mini-batch (last learning cycle in each function call of
fit) can be smaller than
BatchSize. For example, if you specify
BatchSize = 10 and supply 25 observations to
fit, the function uses 10 observations for the first two
learning cycles and 5 observations for the last learning cycle.
The default BatchSize value depends on how you create the model:
If you convert a traditionally trained model to create
Mdl, and you specifySolver="sgd"orSolver="asgd", thenBatchSizeis specified by the corresponding property of the object.If you create
Mdlby callingincrementalOneClassSVMdirectly, the default value is10.
Data Types: single | double
This property is read-only.
Ridge (L2) regularization term strength, specified as a nonnegative scalar.
You cannot specify this parameter unless you specify Solver =
"sgd" or Solver =
"asgd".
If you specify "auto" when creating the model object:
When
Solveris"sgd"or"asgd", the software estimatesLambdaduring the estimation period using a heuristic procedure.When
Solveris"scale-invariant", thenLambda=NaN.
The default Lambda value depends on how you create the model:
If you convert a traditionally trained model to create
Mdl, and you specifySolver="sgd"orSolver="asgd", thenLambdais specified by the corresponding property of the traditionally trained model. If you specify a different solver, the default value isNaN.If you create
Mdlby callingincrementalOneClassSVMdirectly, the default value isNaN.
Data Types: double | single
This property is read-only.
Initial learning rate, specified as "auto" or a positive
scalar. incrementalOneClassSVM stores the LearnRate value
as a numeric scalar.
You cannot specify this parameter when Solver is
"scale-invariant".
The learning rate controls the optimization step size by scaling the objective
subgradient. LearnRate specifies an initial value for the
learning rate, and LearnRateSchedule
determines the learning rate for subsequent learning cycles.
When you specify "auto":
The initial learning rate is
0.7.If
EstimationPeriod>0,fitchanges the rate to1/sqrt(1+max(sum(X.^2,2)))at the end ofEstimationPeriod, whereXis the predictor data collected during the estimation period.
The default LearnRate value depends on how you create the model:
If you create
Mdlby callingincrementalOneClassSVMdirectly, the default value is"auto".Otherwise, the
LearnRatename-value argument of theincrementalLearnerfunction sets this property. The default value of the argument is"auto".
Data Types: single | double | char | string
This property is read-only.
Learning rate schedule, specified as "decaying" or
"constant", where LearnRate specifies
the initial learning rate ɣ0.
incrementalOneClassSVM stores the LearnRateSchedule
value as a character vector.
You cannot specify this parameter unless you specify Solver=
"sgd" or Solver=
"asgd".
| Value | Description |
|---|---|
"constant" | The learning rate is ɣ0 for all learning cycles. |
"decaying" | The learning rate at learning cycle t is
|
The default LearnRateSchedule value depends on how you create
the model:
If you convert a traditionally trained model object to create
Mdl, theLearnRateSchedulename-value argument of theincrementalLearnerfunction sets this property.Otherwise, the default value is
"decaying".
Data Types: char | string
Score Threshold Parameters
This property is read-only.
Flag indicating whether the incremental fitting function fit returns
scores and detects anomalies after training the model, specified as logical
0 (false) or 1
(true).
The incremental model Mdl is warm
(IsWarm becomes true) after the
fit function fits the incremental model to
ScoreWarmupPeriod observations.
You cannot specify IsWarm directly.
If EstimationPeriod >
0, then during the estimation period, fit
does not fit the model or update ScoreThreshold, and
IsWarm is false.
| Value | Description |
|---|---|
1(true) | The incremental model Mdl is warm. Consequently,
fit trains the model and then returns scores and
detects anomalies. |
0(false) | fit trains the model but returns all scores as
NaN and all anomaly indicators as
false. |
Data Types: logical
This property is read-only.
Threshold for the anomaly score used to detect anomalies, specified as a
nonnegative integer. incrementalOneClassSVM detects observations with scores
above the threshold as anomalies.
Note
If EstimationPeriod >
0, then during the estimation period,
fit does not fit the model or update
ScoreThreshold.
The default ScoreThreshold value depends on how you create
the model:
If you convert a traditionally trained model object to create
Mdl, thenScoreThresholdis specified by the corresponding property value of the object.Otherwise, the default value is
0.
You cannot specify ScoreThreshold directly.
Data Types: single | double
This property is read-only.
Warm-up period before score output and anomaly detection (outside the estimation
period, if EstimationPeriod > 0), specified as
a nonnegative integer. The ScoreWarmupPeriod value is the number
of observations to which the incremental model must be fit before the incremental
fit
function returns scores and detects anomalies.
Note
When processing observations during the score warm-up period, the software ignores observations that contain at least one missing value.
You can return scores and detect anomalies during the warm-up period by calling
isanomalydirectly.
The default ScoreWarmupPeriod value depends on how you create
the model:
If you convert a traditionally trained model to create
Mdl, theScoreWarmupPeriodname-value argument of theincrementalLearnerfunction sets this property.Otherwise, the default value is
0.
Data Types: single | double
This property is read-only.
Running window size used to estimate the score threshold
(ScoreThreshold), specified as a positive integer.
The default ScoreWindowSize value depends on how you create
the model:
If you convert a traditionally trained model to create
Mdl, theScoreWindowSizename-value argument of theincrementalLearnerfunction sets this property.Otherwise, the default value is
1000.
Data Types: double
Training Parameters
This property is read-only.
Number of observations processed by the incremental learner to estimate hyperparameters before training, specified as a nonnegative integer.
When processing observations during the estimation period, the software ignores observations that contain at least one missing value.
If
Mdlis prepared for incremental learning (all hyperparameters required for training are specified),incrementalOneClassSVMforcesEstimationPeriodto0.If
Mdlis not prepared for incremental learning,incrementalOneClassSVMsetsEstimationPeriodto1000and estimates the unknown hyperparameters.
For more details, see Estimation Period.
Data Types: single | double
This property is read-only.
Objective function minimization technique, specified as a value in this table.
| Value | Description | Notes |
|---|---|---|
"scale-invariant" | Adaptive scale-invariant solver for incremental learning [1] |
|
"sgd" | Stochastic gradient descent (SGD) [5][2] |
|
"asgd" | Average stochastic gradient descent (ASGD) [6] |
|
Data Types: char | string
This property is read-only.
Objective solver configurations, specified as a structure array. The fields of
SolverOptions depend on the value of Solver.
You can specify the field values using the corresponding name-value arguments when
you create the model object by calling incrementalOneClassSVM directly, or when
you convert a traditionally trained model using the
incrementalLearner function.
You cannot specify SolverOptions directly.
Data Types: struct
Object Functions
Examples
Create a default one-class support vector machine (SVM) model for incremental anomaly detection.
Mdl = incrementalOneClassSVM; Mdl.ScoreWarmupPeriod
ans = 0
Mdl.ContaminationFraction
ans = 0
Mdl is an incrementalOneClassSVM model object. All its properties are read-only. By default, the software sets the score warm-up period to 0 and the anomaly contamination fraction to 0.
Mdl must be fit to data before you can use it to perform any other operations.
Load Data
Load the 1994 census data stored in census1994.mat. The data set consists of demographic data from the US Census Bureau.
load census1994.matincrementalOneClassSVM does not support categorical predictors and does not use observations with missing values. Remove missing values in the data to reduce memory consumption and speed up training. Remove the categorical predictors.
adultdata = rmmissing(adultdata); adultdata = removevars(adultdata,["workClass","education","marital_status", ... "occupation","relationship","race","sex","native_country","salary"]);
Fit Incremental Model
Fit the incremental model Mdl to the data in the adultdata table by using the fit function. Because ScoreWarmupPeriod = 0, fit returns scores and detects anomalies immediately after fitting the model for the first time. To simulate a data stream, fit the model in chunks of 100 observations at a time. At each iteration:
Process 100 observations.
Overwrite the previous incremental model with a new one fitted to the incoming observations.
Store
medianscore, the median score value of the data chunk, to see how it evolves during incremental learning.Store
allscores, the score values for the fitted observations.Store
threshold, the score threshold value for anomalies, to see how it evolves during incremental learning.Store
numAnom, the number of detected anomalies in the data chunk.
n = numel(adultdata(:,1)); numObsPerChunk = 100; nchunk = floor(n/numObsPerChunk); medianscore = zeros(nchunk,1); threshold = zeros(nchunk,1); numAnom = zeros(nchunk,1); allscores = []; % Incremental fitting rng(0,"twister"); % For reproducibility for j = 1:nchunk ibegin = min(n,numObsPerChunk*(j-1) + 1); iend = min(n,numObsPerChunk*j); idx = ibegin:iend; Mdl = fit(Mdl,adultdata(idx,:)); [isanom,scores] = isanomaly(Mdl,adultdata(idx,:)); medianscore(j) = median(scores); allscores = [allscores scores']; numAnom(j) = sum(isanom); threshold(j) = Mdl.ScoreThreshold; end
Mdl is an incrementalOneClassSVM model object trained on all the data in the stream. The fit function fits the model to the data chunk, and the isanomaly function returns the observation scores and the indices of observations in the data chunk with scores above the score threshold value.
Analyze Incremental Model During Training
Plot the anomaly score for every observation.
plot(allscores,".-") xlabel("Observation") ylabel("Score") xlim([0 n])

At each iteration, the software calculates a score value for each observation in the data chunk. A negative score value with large magnitude indicates a normal observation, and a large positive value indicates an anomaly.
To see how the score threshold and median score per data chunk evolve during training, plot them on separate tiles.
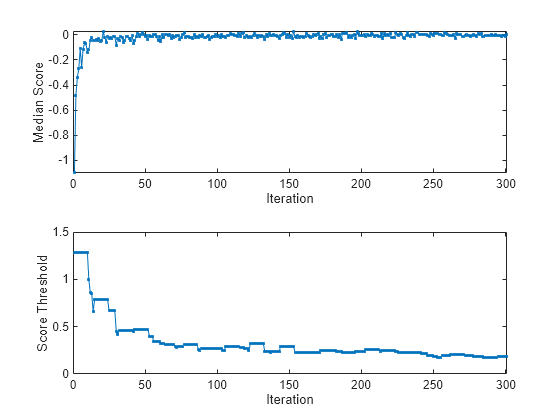
figure tiledlayout(2,1); nexttile plot(medianscore,".-") ylabel("Median Score") xlabel("Iteration") xlim([0 nchunk]) nexttile plot(threshold,".-") ylabel("Score Threshold") xlabel("Iteration") xlim([0 nchunk])
finalScoreThreshold=Mdl.ScoreThreshold
finalScoreThreshold = 0.1799
The median score is negative for the first several iterations, then rapidly approaches zero. The anomaly score threshold immediately rises from its (default) starting value of 0 to 1.3, and then gradually approaches 0.18. Because ContaminationFraction = 0, incrementalOneClassSVM treats all training observations as normal observations, and at each iteration sets the score threshold to the maximum score value in the data chunk.
totalAnomalies = sum(numAnom)
totalAnomalies = 0
No anomalies are detected at any iteration, because ContaminationFraction = 0.
Prepare an incremental one-class SVM model by specifying an anomaly contamination fraction of 0.001, and standardize the data using an initial estimation period of 2000 observations. Specify a score warm-up period of 10,000 observations, during which the fit function updates the score threshold and trains the model but does not return scores or detect anomalies.
Mdl = incrementalOneClassSVM(ContaminationFraction=0.001, ... StandardizeData=true,EstimationPeriod=2000, ... ScoreWarmupPeriod=10000);
Mdl is an incrementalOneClassSVM model object. All its properties are read-only. Mdl must be fit to data before you can use it to perform any other operations.
Load Data
Load the 1994 census data stored in census1994.mat. The data set consists of demographic data from the US Census Bureau.
load census1994.matincrementalOneClassSVM does not support categorical predictors and does not use observations with missing values. Remove missing values in the data to reduce memory consumption and speed up training. Remove the categorical predictors.
adultdata = rmmissing(adultdata); Xtrain = removevars(adultdata,["workClass","education","marital_status", ... "occupation","relationship","race","sex","native_country","salary"]);
Fit Incremental Model and Detect Anomalies
Fit the incremental model Mdl to the data by using the fit function. To simulate a data stream, fit the model in chunks of 100 observations at a time. Because EstimationPeriod = 2000 and ScoreWarmupPeriod = 10000, fit returns scores and detects anomalies only after 120 iterations. At each iteration:
Process 100 observations.
Overwrite the previous incremental model with a new one fitted to the incoming observations.
Store
meanscore, the mean score value of the data chunk, to see how it evolves during incremental learning.Store
threshold, the score threshold value for anomalies, to see how it evolves during incremental learning.Store
numAnom, the number of detected anomalies in the chunk, to see how it evolves during incremental learning.
n = numel(Xtrain(:,1)); numObsPerChunk = 100; nchunk = floor(n/numObsPerChunk); meanscore = zeros(nchunk,1); threshold = zeros(nchunk,1); numAnom = zeros(nchunk,1); % Incremental fitting rng("default"); % For reproducibility for j = 1:nchunk ibegin = min(n,numObsPerChunk*(j-1) + 1); iend = min(n,numObsPerChunk*j); idx = ibegin:iend; [Mdl,tf,scores] = fit(Mdl,Xtrain(idx,:)); meanscore(j) = mean(scores); numAnom(j) = sum(tf); threshold(j) = Mdl.ScoreThreshold; end
Mdl is an incrementalOneClassSVM model object trained on all the data in the stream.
Analyze Incremental Model During Training
To see how the mean score, score threshold, and number of detected anomalies per chunk evolve during training, plot them on separate tiles.
tiledlayout(3,1); nexttile plot(meanscore) ylabel("Mean Score") xlabel("Iteration") xlim([0 nchunk]) xline(Mdl.EstimationPeriod/numObsPerChunk,"r-.") xline((Mdl.EstimationPeriod+Mdl.ScoreWarmupPeriod)/numObsPerChunk,"r") nexttile plot(threshold) ylabel("Score Threshold") xlabel("Iteration") xlim([0 nchunk]) xline(Mdl.EstimationPeriod/numObsPerChunk,"r-.") xline((Mdl.EstimationPeriod+Mdl.ScoreWarmupPeriod)/numObsPerChunk,"r") nexttile plot(numAnom,"+") ylabel("Anomalies") xlabel("Iteration") xlim([0 nchunk]) ylim([0 max(numAnom)+0.2]) xline(Mdl.EstimationPeriod/numObsPerChunk,"r-.") xline((Mdl.EstimationPeriod+Mdl.ScoreWarmupPeriod)/numObsPerChunk,"r")

During the estimation period, fit estimates means and standard deviations using the observations, and does not fit the model or update the score threshold. During the warm-up period, the fit function fits the model and updates the score threshold, but returns all scores as NaN and all anomaly values as false. After the warm-up period, fit returns the observation scores and the indices of observations with scores above the score threshold value. A negative score value with large magnitude indicates a normal observation, and a large positive value indicates an anomaly.
totalAnomalies=sum(numAnom)
totalAnomalies = 18
anomfrac= totalAnomalies/(n-Mdl.EstimationPeriod-Mdl.ScoreWarmupPeriod)
anomfrac = 9.9108e-04
The software detected 18 anomalies after the warm-up and estimation periods. The contamination fraction after the estimation and warm-up periods is approximately 0.001.
More About
Incremental learning, or online learning, is a branch of machine learning concerned with processing incoming data from a data stream, possibly given little to no knowledge of the distribution of the predictor variables, aspects of the prediction or objective function (including tuning parameter values), or whether the observations contain anomalies. Incremental learning differs from traditional machine learning, where enough data is available to fit to a model, perform cross-validation to tune hyperparameters, and infer the predictor distribution.
Anomaly detection is used to identify unexpected events and departures from normal behavior. In situations where the full data set is not immediately available, or new data is arriving, you can use incremental learning for anomaly detection to incrementally train a model so it adjusts to the characteristics of the incoming data.
Given incoming observations, an incremental learning model for anomaly detection does the following:
Computes anomaly scores
Updates the anomaly score threshold
Detects data points above the score threshold as anomalies
Fits the model to the incoming observations
For more information, see Incremental Anomaly Detection with MATLAB.
The adaptive scale-invariant solver for incremental learning, introduced in [1], is a gradient-descent-based objective solver for training linear predictive models. The solver is hyperparameter free, insensitive to differences in predictor variable scales, and does not require prior knowledge of the distribution of the predictor variables. These characteristics make it well suited to incremental learning.
The incremental fitting function fit uses the more
aggressive ScInOL2 version of the algorithm to train binary learners. The function always
shuffles an incoming batch of data before fitting the model.
Random feature expansion, such as Random Kitchen Sinks [4] or Fastfood [3], is a scheme to approximate Gaussian kernels of the kernel classification algorithm to use for big data in a computationally efficient way. Random feature expansion is more practical for big data applications that have large training sets, but can also be applied to smaller data sets that fit in memory.
The kernel classification algorithm searches for an optimal hyperplane that separates the data into two classes after mapping features into a high-dimensional space. Nonlinear features that are not linearly separable in a low-dimensional space can be separable in the expanded high-dimensional space. All the calculations for hyperplane classification use only dot products. You can obtain a nonlinear classification model by replacing the dot product x1x2' with the nonlinear kernel function , where xi is the ith observation (row vector) and φ(xi) is a transformation that maps xi to a high-dimensional space (called the “kernel trick”). However, evaluating G(x1,x2) (Gram matrix) for each pair of observations is computationally expensive for a large data set (large n).
The random feature expansion scheme finds a random transformation so that its dot product approximates the Gaussian kernel. That is,
where T(x) maps x in to a high-dimensional space (). The Random Kitchen Sinks scheme uses the random transformation
where is a sample drawn from and σ is a kernel scale. This scheme requires O(mp) computation and storage.
The Fastfood scheme introduces another random
basis V instead of Z using Hadamard matrices combined
with Gaussian scaling matrices. This random basis reduces the computation cost to O(mlogp) and reduces storage to O(m).
You can specify values for m and
σ by setting NumExpansionDimensions and
KernelScale, respectively, of incrementalOneClassSVM.
The incrementalOneClassSVM function uses the
Fastfood scheme for random feature expansion, and uses linear classification to train a
one-class Gaussian kernel classification model.
Algorithms
During the estimation period, the incremental fitting function fit uses the
first incoming EstimationPeriod observations to estimate (tune)
hyperparameters required for incremental training. Estimation occurs only when
EstimationPeriod is positive. This table describes the
hyperparameters and when they are estimated, or tuned.
| Hyperparameter | Model Property | Usage | Conditions |
|---|---|---|---|
| Predictor means and standard deviations |
| Standardize predictor data | The hyperparameters are estimated when both of these conditions apply:
|
| Learning rate | LearnRate field of SolverOptions | Adjust the solver step size | The hyperparameter is estimated when both of these conditions apply:
|
| Kernel scale parameter | KernelScale | Set a kernel scale parameter value for random feature expansion | The hyperparameter is estimated when you set the
KernelScale to "auto". |
During the estimation period,
fit does not fit the model. At the end of the estimation period,
the function updates the properties that store the hyperparameters.
If incremental learning functions are configured to standardize predictor variables,
they do so using the means and standard deviations stored in the Mu and
Sigma properties of the incremental learning model
Mdl.
When you set
StandardizeData=trueand a positive estimation period (seeEstimationPeriod), andMdl.MuandMdl.Sigmaare empty, the incremental fit function estimates means and standard deviations using the estimation period observations.When you set
StandardizeData="auto"(the default), the following conditions apply:If you create
incrementalOneClassSVMby converting a traditionally trained one-class SVM model (OneClassSVM), and theMuandSigmaproperties of the model being converted are empty arrays[], incremental learning functions do not standardize predictor variables. If theMuandSigmaproperties of the model being converted are nonempty, incremental learning functions standardize the predictor variables using the specified means and standard deviations. The incremental fitting function does not estimate new means and standard deviations, regardless of the length of the estimation period.If you do not convert a traditionally trained model, the incremental fitting function standardizes the predictor variables only when you specify an SGD solver (see
Solver) and a positive estimation period (seeEstimationPeriod).
When the incremental fitting function estimates predictor means and standard deviations, the function computes weighted means and weighted standard deviations using the estimation period observations. Specifically, the function standardizes predictor j (xj) using
xj is predictor j, and xjk is observation k of predictor j in the estimation period.
wj is observation weight j.
The observation weights wj are all equal to one and cannot be specified.
References
[1] Kempka, Michał, Wojciech Kotłowski, and Manfred K. Warmuth. "Adaptive Scale-Invariant Online Algorithms for Learning Linear Models." Preprint, submitted February 10, 2019. https://arxiv.org/abs/1902.07528.
[2] Langford, J., L. Li, and T. Zhang. “Sparse Online Learning Via Truncated Gradient.” J. Mach. Learn. Res., Vol. 10, 2009, pp. 777–801.
[3] Le, Q., T. Sarlós, and A. Smola. “Fastfood — Approximating Kernel Expansions in Loglinear Time.” Proceedings of the 30th International Conference on Machine Learning. Vol. 28, No. 3, 2013, pp. 244–252.
[4] Rahimi, A., and B. Recht. “Random Features for Large-Scale Kernel Machines.” Advances in Neural Information Processing Systems. Vol. 20, 2008, pp. 1177–1184.
[5] Shalev-Shwartz, S., Y. Singer, and N. Srebro. “Pegasos: Primal Estimated Sub-Gradient Solver for SVM.” Proceedings of the 24th International Conference on Machine Learning, ICML ’07, 2007, pp. 807–814.
[6] Xu, Wei. “Towards Optimal One Pass Large Scale Learning with Averaged Stochastic Gradient Descent.” CoRR, abs/1107.2490, 2011.
Version History
Introduced in R2023b
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
웹사이트 선택
번역된 콘텐츠를 보고 지역별 이벤트와 혜택을 살펴보려면 웹사이트를 선택하십시오. 현재 계신 지역에 따라 다음 웹사이트를 권장합니다:
또한 다음 목록에서 웹사이트를 선택하실 수도 있습니다.
사이트 성능 최적화 방법
최고의 사이트 성능을 위해 중국 사이트(중국어 또는 영어)를 선택하십시오. 현재 계신 지역에서는 다른 국가의 MathWorks 사이트 방문이 최적화되지 않았습니다.
미주
- América Latina (Español)
- Canada (English)
- United States (English)
유럽
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)