피크 분석
이 예제에서는 기본적인 피크 분석을 수행하는 방법을 보여줍니다. 이는 다음과 같은 문제의 해결에 도움이 됩니다. 신호의 피크는 어떻게 찾습니까? 피크 간의 거리는 어떻게 측정합니까? 추세의 영향을 받는 신호의 피크 진폭은 어떻게 측정합니까? 잡음이 있는 신호의 피크는 어떻게 찾습니까? 국소 최솟값은 어떻게 구합니까?
최댓값 또는 피크 찾기
취리히 흑점 상대 수(Zurich Sunspot Relative Number)는 태양 흑점의 개수와 크기를 모두 측정합니다. findpeaks 함수를 사용하여 피크의 위치와 값을 찾을 수 있습니다.
load sunspot.dat year = sunspot(:,1); relNums = sunspot(:,2); findpeaks(relNums,year) xlabel("Year") ylabel("Sunspot Number") title("Find All Peaks")

위에 나와 있는 플롯은 300년 동안 표로 만들어 온 태양 흑점의 개수를 보여주고 검출된 피크를 표시합니다. 다음 섹션에서는 이러한 피크 간 거리를 측정하는 방법을 보여줍니다.
피크 간 거리 측정하기
신호의 피크가 규칙적인 간격으로 나타나는 것처럼 보입니다. 그러나, 일부 피크는 서로 매우 가깝게 있습니다. MinPeakProminence 이름-값 인수를 사용하여 이들 피크를 필터링할 수 있습니다. 피크의 양쪽 방향으로 이전 값보다 더 큰 값이 나타나기 전까지 상대흑점수가 최소 40개 이상 감소하는 피크를 고려합니다.
findpeaks(relNums,year,MinPeakProminence=40) xlabel("Year") ylabel("Sunspot Number") title("Find Prominent Peaks")

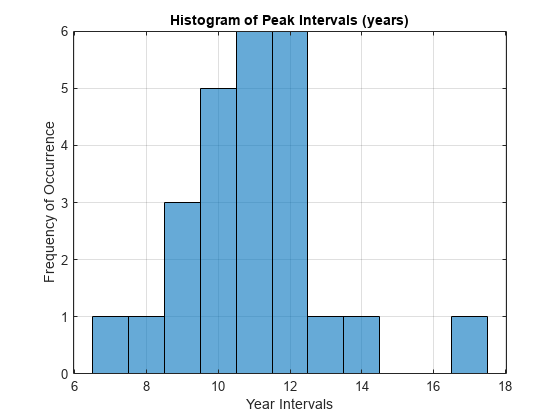
다음 히스토그램에서는 수년 동안의 피크 간격 분포를 보여줍니다.
figure [pks,locs] = findpeaks(relNums,year,MinPeakProminence=40); peakInterval = diff(locs); histogram(peakInterval) grid on xlabel("Year Intervals") ylabel("Frequency of Occurrence") title("Histogram of Peak Intervals (years)")
AverageDistance_Peaks = mean(diff(locs))
AverageDistance_Peaks = 10.9600
이 분포는 대부분의 피크 간격이 10년에서 12년 사이에 있음을 보여주며, 이는 신호가 주기적인 특성을 가지고 있음을 나타냅니다. 또한, 피크 간 10.96년의 평균 간격은 11년이라고 알려진 주기적인 태양 흑점의 활동과 일치합니다.
잘려진 신호나 포화된 신호에서 피크 찾기
평탄한 피크를 피크로 간주하거나 제외하고자 할 수 있습니다. 후자의 경우, 한 피크와 바로 근방 피크 간의 진폭 차이로 정의되는 최소 편위를 Threshold 이름-값 인수를 사용하여 지정합니다.
load clippedpeaks.mat figure % Show all peaks in the first plot tiledlayout("flow") ax(1) = nexttile; findpeaks(saturatedData) xlabel("Samples") ylabel("Amplitude") title("Detecting Saturated Peaks") % Specify a minimum excursion in the second plot ax(2) = nexttile; findpeaks(saturatedData,Threshold=5) xlabel("Samples") ylabel("Amplitude") title("Filtering Out Saturated Peaks") % link and zoom in to show the changes linkaxes(ax(1:2),"xy") axis(ax,[50 70 0 250])

첫 번째 서브플롯은 평탄한 피크의 경우 상승하는 경계값이 피크로 검출된다는 것을 보여줍니다. 두 번째 서브플롯은 임계값을 지정하면 평탄한 피크를 제외하는 데 도움이 될 수 있음을 보여줍니다.
피크의 진폭 측정하기
이 예제에서는 심전도(ECG) 신호의 피크 분석을 보여줍니다. 심전도는 시간에 따른 심장의 전기적 활성을 측정한 것입니다. 이 신호는 피부에 부착된 전극을 통해 측정되며 움직임 아티팩트로 인한 잡음과 전원 간섭 등의 방해에 민감합니다.
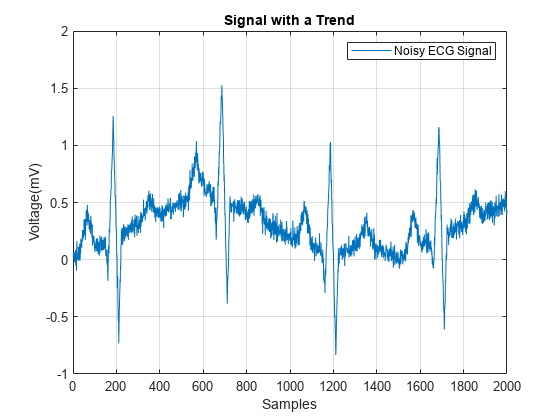
load noisyecg.mat t = 1:length(noisyECG_withTrend); figure plot(t,noisyECG_withTrend) title("Signal with Trend") xlabel("Samples"); ylabel("Voltage (mV)") legend("Noisy ECG Signal") grid on
데이터 추세 제거(디트렌딩)
위에 나와 있는 신호는 기준선이 이동되었기 때문에 실제 진폭을 나타내지 못하고 있습니다. 추세를 제거하기 위해 저차 다항식을 신호에 피팅하고 이 다항식을 사용하여 신호에 대한 추세 제거를 수행합니다.
ECG_data = detrend(noisyECG_withTrend,6); figure plot(t,ECG_data) grid on ax = axis; axis([ax(1:2) -1.2 1.2]) title("Detrended ECG Signal") xlabel("Samples") ylabel("Voltage (mV)") legend("Detrended ECG Signal")

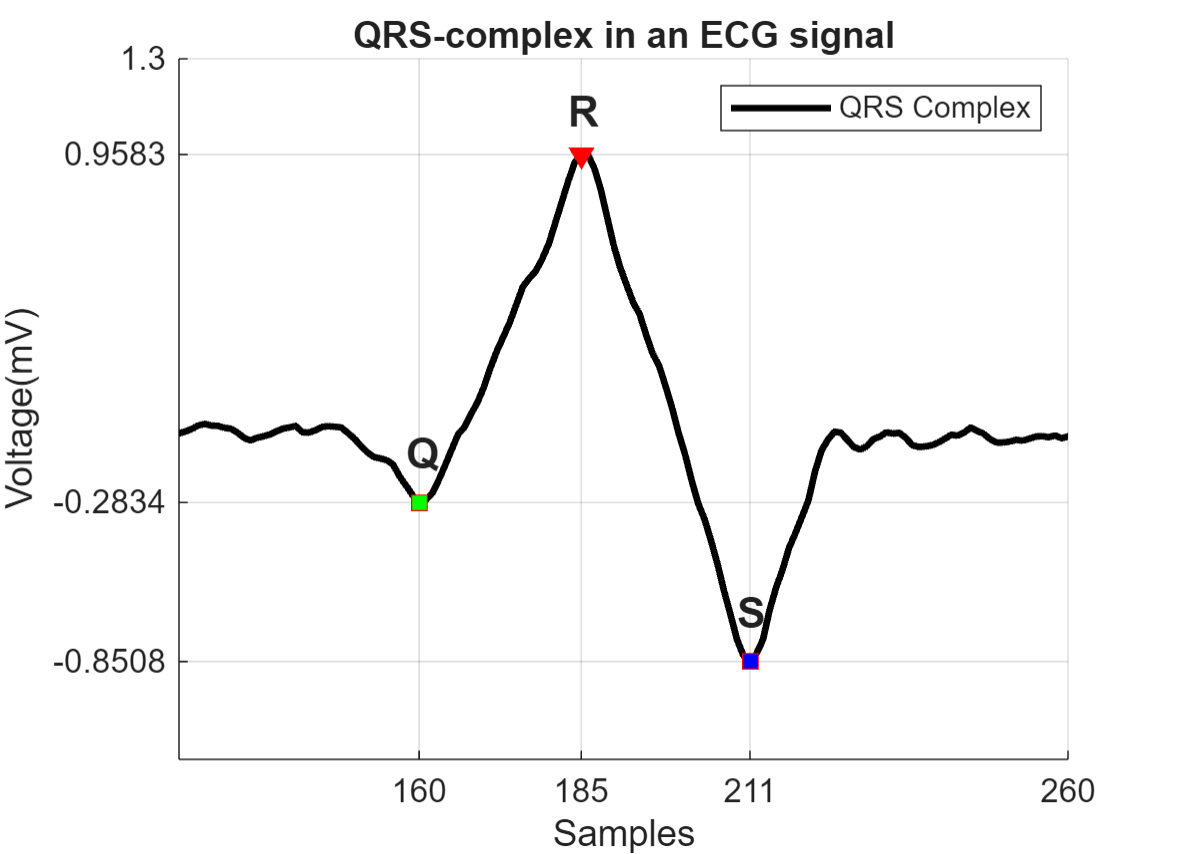
추세 제거를 수행한 후 심전도 신호에서 가장 두드러지게 반복 출현하는 피크인 QRS 복합파를 찾습니다. QRS 복합파는 사람 심장의 우심실과 좌심실의 탈분극에 해당합니다. 이를 사용하여 환자의 심장박동수를 확인하거나 심장 기능의 이상을 예측할 수 있습니다. 다음 그림은 심전도 신호에서 검출되는 QRS 복합파의 파형을 보여줍니다.
임계값을 지정하여 관심 있는 피크 찾기
QRS 복합파는 세 가지 주요 성분인 Q파, R파, S파로 구성됩니다. R파는 피크의 임계값을 0.5mV보다 높게 지정하여 검출할 수 있습니다. 200개 이상의 샘플을 사용하면 R파가 분리되는 것을 알 수 있습니다. 이 정보를 활용하여 MinPeakDistance 이름-값 인수를 지정하여 원치 않는 피크를 제거합니다.
[~,locs_Rwave] = findpeaks(ECG_data, ...
MinPeakHeight=0.5,MinPeakDistance=200);S파 검출을 위해서는 신호에서 국소 최솟값을 구하고 임계값을 적절하게 적용해야 합니다.
신호에서 국소 최솟값 찾기
국소 최솟값은 원래 신호를 반전한 버전에서 피크를 찾는 방식으로 검출할 수 있습니다.
ECG_inverted = -ECG_data;
[~,locs_Swave] = findpeaks(ECG_inverted, ...
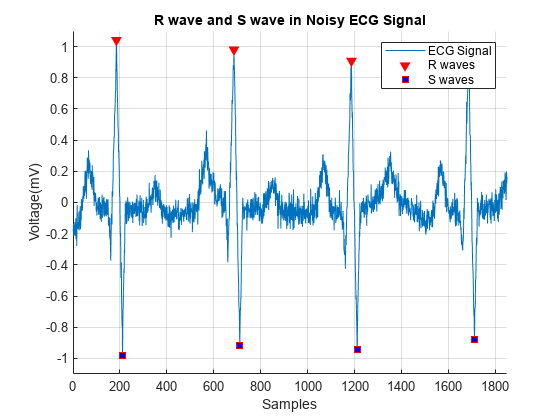
MinPeakHeight=0.5,MinPeakDistance=200);다음 플롯에서는 신호에서 검출한 R파와 S파를 보여줍니다.
figure hold on plot(t,ECG_data) plot(locs_Rwave,ECG_data(locs_Rwave),"v") plot(locs_Swave,ECG_data(locs_Swave),"*") axis([0 1850 -1.1 1.1]) grid on legend("ECG Signal","R wave","S wave") xlabel("Samples") ylabel("Voltage (mV)") title("R wave and S wave in Noisy ECG Signal")
다음으로, Q파의 위치를 확인해 봅니다. Q파를 찾기 위해 피크의 임계값을 지정하면 Q파가 잡음에 묻혀 있기 때문에 원치 않는 피크가 검출됩니다. 먼저 신호를 필터링한 후 피크를 찾습니다. 사비츠키-골레이 필터링(Savitzky-Golay Filtering)을 사용하여 신호에서 잡음을 제거합니다.
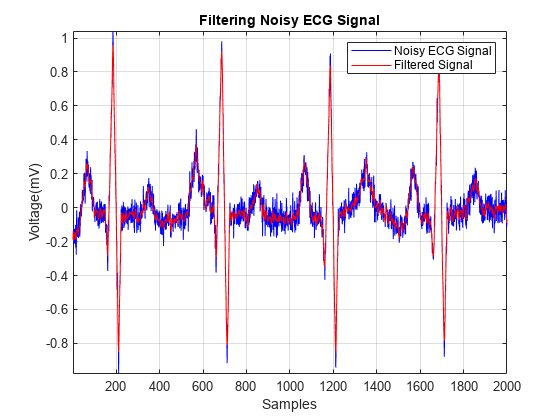
smoothECG = sgolayfilt(ECG_data,7,21); figure plot(t,ECG_data,t,smoothECG) grid on axis tight xlabel("Samples") ylabel("Voltage (mV)") legend("Noisy ECG Signal","Filtered Signal") title("Filtering Noisy ECG Signal")
매끄러운 신호에서 피크 검출을 수행하고 논리형 인덱싱을 사용하여 Q파의 위치를 찾습니다.
[~,min_locs] = findpeaks(-smoothECG,MinPeakDistance=40); % Peaks between -0.2 mV and -0.5 mV locs_Qwave = min_locs(smoothECG(min_locs) > -0.5 ... & smoothECG(min_locs) < -0.2); figure hold on plot(t,smoothECG); plot(locs_Rwave,smoothECG(locs_Rwave),"v") plot(locs_Swave,smoothECG(locs_Swave),"*") plot(locs_Qwave,smoothECG(locs_Qwave),"o") hold off grid on title("Thresholding Peaks in Signal") xlabel("Samples") ylabel("Voltage(mV)") ax = axis; axis([0 1850 -1.1 1.1]) legend("Smooth ECG signal","R wave","S wave","Q wave")
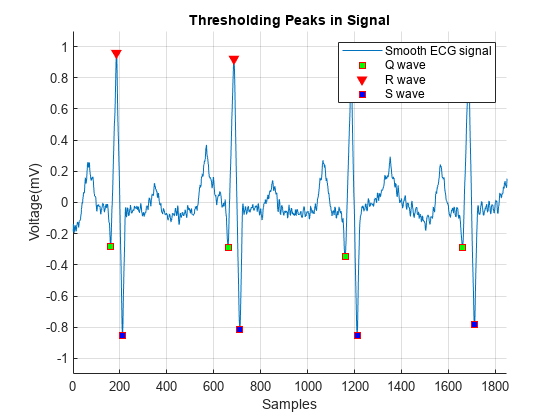
위 그림은 잡음이 있는 심전도 신호에서 성공적으로 검출한 QRS 복합파를 보여줍니다.
잡음이 있는 신호와 매끄러운 신호 사이의 오차
원시 신호의 QRS 복합파와 추세 제거되고 필터링된 신호의 QRS 복합파 사이의 평균 차이를 확인해 봅니다.
% Values of the Extrema [val_Qwave, val_Rwave, val_Swave] = deal(smoothECG(locs_Qwave), ... smoothECG(locs_Rwave), smoothECG(locs_Swave)); meanError_Qwave = mean((noisyECG_withTrend(locs_Qwave) - val_Qwave))
meanError_Qwave = 0.2771
meanError_Rwave = mean((noisyECG_withTrend(locs_Rwave) - val_Rwave))
meanError_Rwave = 0.3476
meanError_Swave = mean((noisyECG_withTrend(locs_Swave) - val_Swave))
meanError_Swave = 0.1844
이는 효율적인 피크 분석을 위해서는 잡음이 있는 신호를 반드시 추세 제거해야 한다는 것을 보여줍니다.
피크 특성
피크의 중요한 특성으로는 상승 시간, 하강 시간, 상승 레벨, 하강 레벨이 있습니다. 이러한 특성은 심전도 신호의 QRS 복합파 각각에 대해 계산됩니다. 이러한 특성의 평균값은 아래 그림에 표시되어 있습니다.
avg_riseTime = mean(locs_Rwave-locs_Qwave); % Average Rise time avg_fallTime = mean(locs_Swave-locs_Rwave); % Average Fall time avg_riseLevel = mean(val_Rwave-val_Qwave); % Average Rise Level avg_fallLevel = mean(val_Rwave-val_Swave); % Average Fall Level helperPeakAnalysisPlot(t,smoothECG, ... locs_Qwave,locs_Rwave,locs_Swave, ... val_Qwave,val_Rwave,val_Swave, ... avg_riseTime,avg_fallTime, ... avg_riseLevel,avg_fallLevel)
