similarityDistance
Syntax
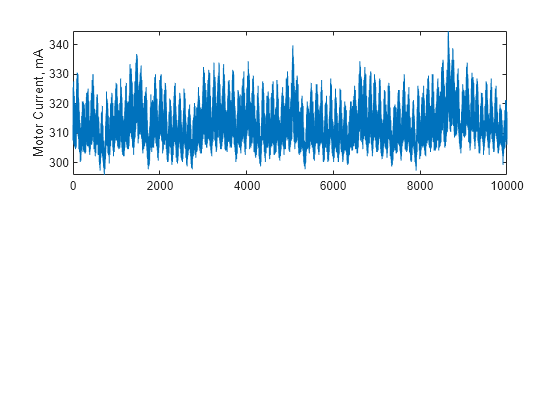
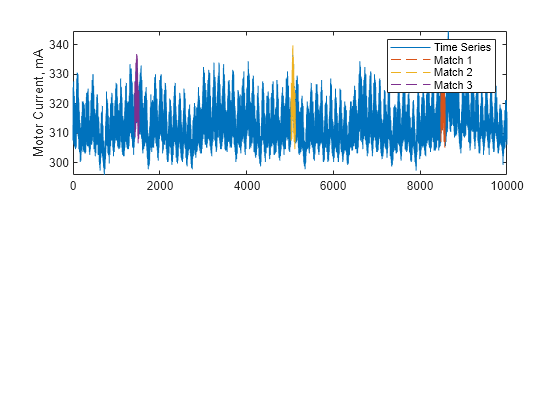
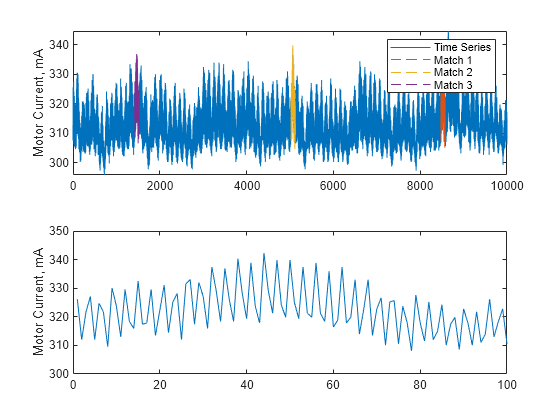
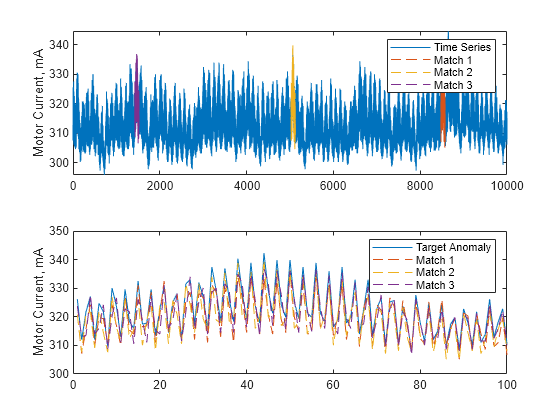
Description
Examples
Input Arguments
Output Arguments
References
[1] Abdullah Mueen, Sheng Zhong, Yan Zhu, Michael Yeh, Kaveh Kamgar, Krishnamurthy Viswanathan, Chetan Kumar Gupta, and Eamonn Keogh, The Fastest Similarity Search Algorithm for Time Series Subsequences Under Euclidean Distance, 2022. https://www.cs.unm.edu/%7Emueen/FastestSimilaritySearch.html
Extended Capabilities
Version History
Introduced in R2024b