shapeFeatures
Description
Examples
Load an X-ray image into the workspace as a medicalImage object. Visualize the image.
data = medicalImage("forearmXrayImage1.dcm");
I = data.Pixels;
figure
imshow(I,[])Draw two regions of interest (ROI) in the X-ray image. Create masks from the ROIs.
roi1 = drawassisted(Color="g"); roi2 = drawassisted(Color="r");
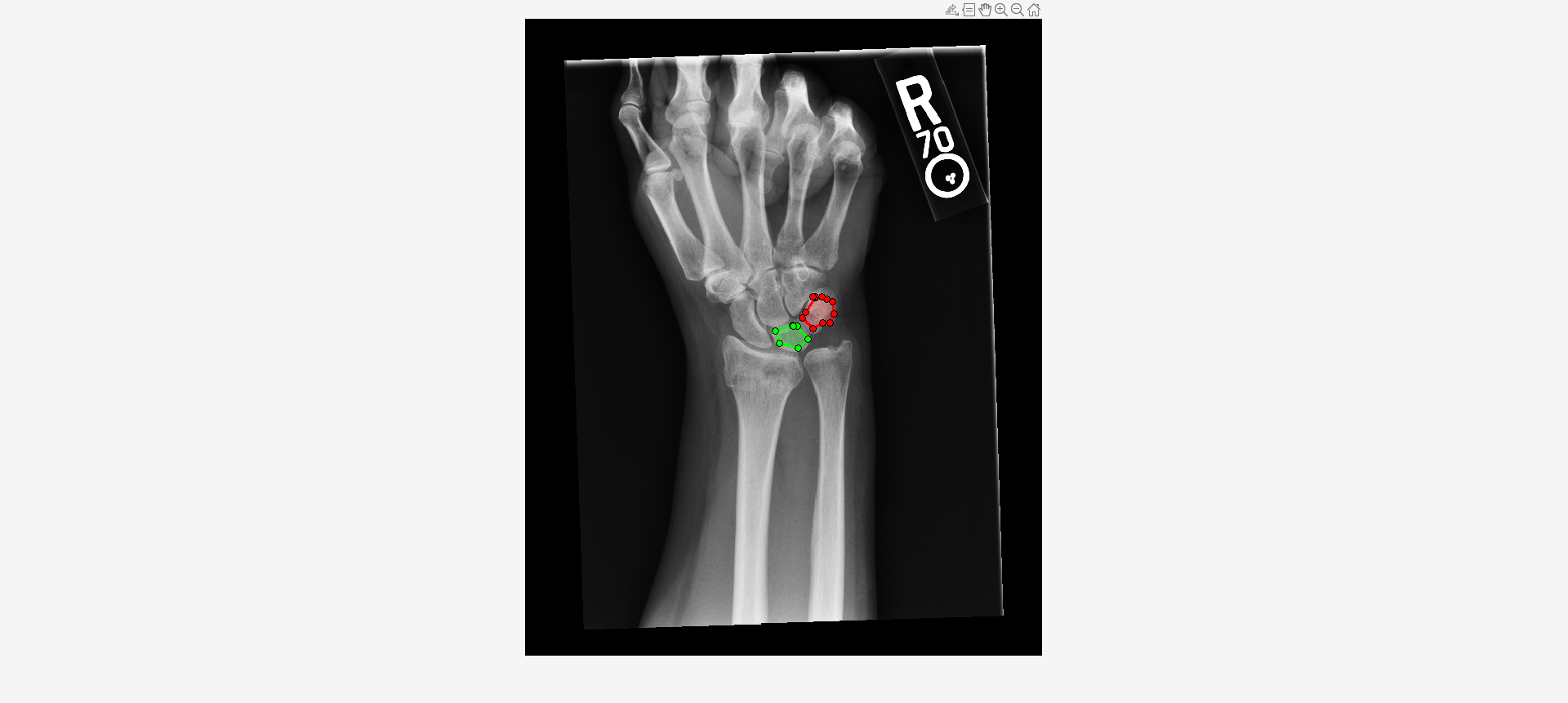
mask1 = createMask(roi1,I); mask2 = createMask(roi2,I);
Create an ROI label matrix, using different labels for the two ROIs. Create a medicalImage object of the ROI label data.
mask = zeros(size(I));
mask(mask1) = 1;
mask(mask2) = 2;
info = dicominfo("forearmXrayImage1.dcm");
roi = medicalImage(mask,info);Create a radiomics object from the X-ray image data and ROI label data.
R = radiomics(data,roi)
R =
radiomics with properties:
Data: [1×1 medicalImage]
ROILabel: [1×1 medicalImage]
Resample: 0
Resegment: 1
Discretize: 1
DiscretizeIVH: 1
ResampledVoxelSpacing: []
DataResampleMethod: []
MaskResampleMethod: []
ResegmentationRange: []
ExcludeOutliers: 1
DiscreteBinSizeOrBinNumber: []
DiscreteMethod: 'FixedBinNumber'
DiscreteIVHBinSizeOrBinNumber: []
DiscreteIVHMethod: 'FixedBinNumber'
Compute shape features for both ROIs.
S = shapeFeatures(R)
S=2×24 table
LabelID VolumeMesh2D VolumeVoxelCount2D SurfaceAreaMesh2D SurfaceVolumeRatio2D Compactness1_2D Compactness2_2D SphericalDisproportion2D Sphericity2D Asphericity2D CentreOfMassShift2D Maximum3dDiameter2D MajorAxisLength2D MinorAxisLength2D LeastAxisLength2D Elongation2D Flatness2D VolumeDensityAABB_2D AreaDensityAABB_2D VolumeDensityAEE_2D AreaDensityAEE_2D VolumeDensityConvexHull2D AreaDensityConvexHull2D IntegratedIntensity2D
_______ ____________ __________________ _________________ ____________________ _______________ _______________ ________________________ ____________ _____________ ___________________ ___________________ _________________ _________________ _________________ ____________ __________ ____________________ __________________ ___________________ _________________ _________________________ _______________________ _____________________
"1" 54 54.427 135.84 2.5156 0.019243 0.13156 1.9662 0.5086 0.96618 0.18567 11.327 10.188 6.924 0 0.67962 0 0.63176 0.65158 NaN 0.91886 0.98182 0.99044 1.2498e+05
"2" 74 74.85 181.6 2.454 0.017061 0.10342 2.1304 0.46939 1.1304 0.24847 11.563 11.187 8.7535 0 0.78246 0 0.64598 0.66783 NaN 0.83862 0.93671 0.9552 1.8045e+05
Import a computed tomography (CT) image volume and the corresponding ROI mask volume from the IBSI validation data set [1][2][3] as medicalVolume objects.
unzip("CTImageMaskNIfTI.zip") data = medicalVolume("CT_image.nii.gz"); roi = medicalVolume("CT_mask.nii.gz");
Visualize a slice of the CT image volume and the corresponding ROI.
figure
imshowpair(data.Voxels(:,:,20),roi.Voxels(:,:,20),"montage")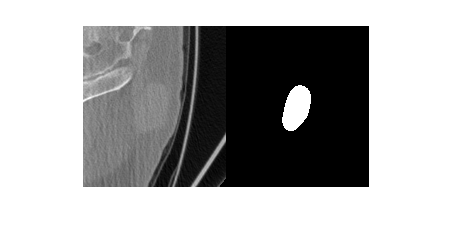
Create a radiomics object, using the CT image volume and ROI mask volume, with default preprocessing options.
R = radiomics(data,roi)
R =
radiomics with properties:
Data: [1×1 medicalVolume]
ROILabel: [1×1 medicalVolume]
Resample: 1
Resegment: 1
Discretize: 1
DiscretizeIVH: 1
ResampledVoxelSpacing: 1
DataResampleMethod: 'linear'
MaskResampleMethod: 'linear'
ResegmentationRange: []
ExcludeOutliers: 1
DiscreteBinSizeOrBinNumber: []
DiscreteMethod: 'FixedBinNumber'
DiscreteIVHBinSizeOrBinNumber: []
DiscreteIVHMethod: 'FixedBinNumber'
Compute the shape features of the ROI in both the 2-D resampled and 3-D resampled CT image volumes.
S = shapeFeatures(R,Type="all",SubType="all")
S=1×51 table
LabelID VolumeMesh2D VolumeVoxelCount2D SurfaceAreaMesh2D SurfaceVolumeRatio2D Compactness1_2D Compactness2_2D SphericalDisproportion2D Sphericity2D Asphericity2D CentreOfMassShift2D Maximum3dDiameter2D MajorAxisLength2D MinorAxisLength2D LeastAxisLength2D Elongation2D Flatness2D VolumeDensityAABB_2D AreaDensityAABB_2D VolumeDensityAEE_2D AreaDensityAEE_2D VolumeDensityConvexHull2D AreaDensityConvexHull2D IntegratedIntensity2D MoransIIndex2D GearysCMeasure2D VolumeMesh3D VolumeVoxelCount3D SurfaceAreaMesh3D SurfaceVolumeRatio3D Compactness1_3D Compactness2_3D SphericalDisproportion3D Sphericity3D Asphericity3D CentreOfMassShift3D Maximum3dDiameter3D MajorAxisLength3D MinorAxisLength3D LeastAxisLength3D Elongation3D Flatness3D VolumeDensityAABB_3D AreaDensityAABB_3D VolumeDensityAEE_3D AreaDensityAEE_3D VolumeDensityConvexHull3D AreaDensityConvexHull3D IntegratedIntensity3D MoransIIndex3D GearysCMeasure3D
_______ ____________ __________________ _________________ ____________________ _______________ _______________ ________________________ ____________ _____________ ___________________ ___________________ _________________ _________________ _________________ ____________ __________ ____________________ __________________ ___________________ _________________ _________________________ _______________________ _____________________ ______________ ________________ ____________ __________________ _________________ ____________________ _______________ _______________ ________________________ ____________ _____________ ___________________ ___________________ _________________ _________________ _________________ ____________ __________ ____________________ __________________ ___________________ _________________ _________________________ _______________________ _____________________ ______________ ________________
"1" 52504 52637 8411.4 0.1602 0.038399 0.52389 1.2405 0.80615 0.24047 0.91477 56.989 50.304 44 33.402 0.87467 0.664 0.47158 0.59688 1.3563 1.4179 0.90243 1.1059 3.1173e+06 0.013525 0.97867 52923 52975 8425 0.15919 0.038611 0.5297 1.2359 0.80911 0.23592 0.8888 57.359 50.405 44.1 33.518 0.8749 0.66497 0.46924 0.59248 1.3566 1.4135 0.89714 1.0994 3.1069e+06 0.016362 0.96493
[1] Vallières, Martin, Carolyn R. Freeman, Sonia R. Skamene, and Issam El Naqa. “A Radiomics Model from Joint FDG-PET and MRI Texture Features for the Prediction of Lung Metastases in Soft-Tissue Sarcomas of the Extremities.” The Cancer Imaging Archive, 2015. https://doi.org/10.7937/K9/TCIA.2015.7GO2GSKS.
[2] Vallières, M, C R Freeman, S R Skamene, and I El Naqa. “A Radiomics Model from Joint FDG-PET and MRI Texture Features for the Prediction of Lung Metastases in Soft-Tissue Sarcomas of the Extremities.” Physics in Medicine and Biology 60, no. 14 (July 7, 2015): 5471–96. https://doi.org/10.1088/0031-9155/60/14/5471.
[3] Clark, Kenneth, Bruce Vendt, Kirk Smith, John Freymann, Justin Kirby, Paul Koppel, Stephen Moore, et al. “The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository.” Journal of Digital Imaging 26, no. 6 (December 2013): 1045–57. https://doi.org/10.1007/s10278-013-9622-7.
Input Arguments
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Example: shapeFeatures(R,SubType="2D") computes the shape features on
2-D resampled data.
Category of shape features to compute, specified as one of these options.
"basic""all"
If you specify "all", the function computes advanced
features in addition to basic features. For more information on which specific shape
features each category includes, see IBSI Standard and Radiomics Function Feature Correspondences.
Data Types: char | string
Resampling from which to compute shape features, specified as one of these options.
"2D"— Computes features from the 2-D resampled data. This is the default option for 2-D data, when theDataandROILabelproperties of theradiomicsobjectRare 2-D matrices ormedicalImageobjects."3D"— Computes features from the 3-D resampled data. This is the default option for 3-D data, when theDataandROILabelproperties of theradiomicsobjectRare 3-D arrays ormedicalVolumeobjects. 3-D resampling is not applicable for 2-D data."all"— Computes features for all applicable options.
For 3-D data, when you perform 3-D resampling of the volume, the function makes the voxel spacing along all three spatial dimensions isotropic. However, when you perform 2-D resampling of the volume, the function makes the voxel spacing along only the x- and y-dimensions isotropic, while retaining the voxel spacing of the input volume along the z-dimension.
Data Types: char | string
Output Arguments
Shape features, returned as a table. The first column in S is
LabelID. The subsequent columns are the shape features. Each row of
the table corresponds to an ROI. For more details on which shape features are computed
in each Type and SubType, see IBSI Standard and Radiomics Function Feature Correspondences.
Version History
Introduced in R2023bCompute shape features for multiple regions of interest (ROIs) in a medical image or medical volume.
Compute two new shape features, MoransIIndex and
GearyCMeasure, in the region of interest of a medical volume. Specify
whether you want to compute basic shape features or all shape features using the new
name-value argument Type.
Compute two new shape features, maximum3dDiameter and
AreaDensityAEE, in the region of interest of a medical volume.
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
웹사이트 선택
번역된 콘텐츠를 보고 지역별 이벤트와 혜택을 살펴보려면 웹사이트를 선택하십시오. 현재 계신 지역에 따라 다음 웹사이트를 권장합니다:
또한 다음 목록에서 웹사이트를 선택하실 수도 있습니다.
사이트 성능 최적화 방법
최고의 사이트 성능을 위해 중국 사이트(중국어 또는 영어)를 선택하십시오. 현재 계신 지역에서는 다른 국가의 MathWorks 사이트 방문이 최적화되지 않았습니다.
미주
- América Latina (Español)
- Canada (English)
- United States (English)
유럽
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)