genfis
Generate fuzzy inference system object from data
Description
fis = genfis(inputData,outputData)
fis = genfis(inputData,outputData,options)options. You can generate fuzzy systems using grid
partitioning, subtractive clustering, or fuzzy c-means (FCM) clustering.
Examples
Define training data.
inputData = [rand(10,1) 10*rand(10,1)-5]; outputData = rand(10,1);
Generate a fuzzy inference system.
fis = genfis(inputData,outputData);
The generated system, fis, is created using grid partitioning with default options.
Define training data.
inputData = [rand(10,1) 10*rand(10,1)-5]; outputData = rand(10,1);
Create a default genfisOptions option set for grid partitioning.
opt = genfisOptions("GridPartition");Specify the following input membership functions for the generated FIS:
3Gaussian membership functions for the first input variable5triangular membership functions for the second input variable
opt.NumMembershipFunctions = [3 5]; opt.InputMembershipFunctionType = ["gaussmf" "trimf"];
Generate the FIS.
fis = genfis(inputData,outputData,opt);
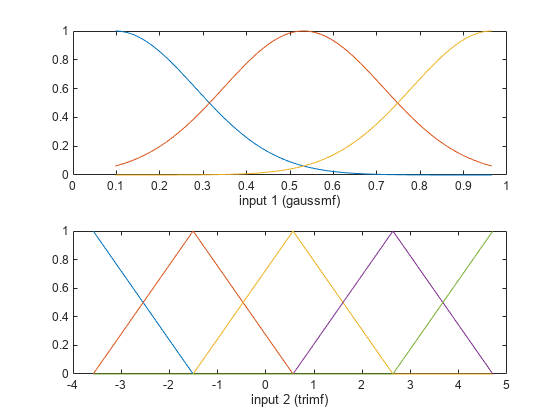
Plot the input membership functions. Each input variable has the specified number and type of input membership functions, evenly distributed over their input range.
[x,mf] = plotmf(fis,"input",1); subplot(2,1,1) plot(x,mf) xlabel("input 1 (gaussmf)") [x,mf] = plotmf(fis,"input",2); subplot(2,1,2) plot(x,mf) xlabel("input 2 (trimf)")
Obtain input and output training data.
load clusterDemo.dat
inputData = clusterDemo(:,1:2);
outputData = clusterDemo(:,3);Create a genfisOptions option set and specify the range of influence for each data dimension. Specify 0.5 and 0.25 as the range of influence for the first and second input variables. Specify 0.3 as the range of influence for the output data.
opt = genfisOptions("SubtractiveClustering", ... ClusterInfluenceRange=[0.5 0.25 0.3]);
Generate the FIS.
fis = genfis(inputData,outputData,opt);
The generated FIS contains one rule for each cluster.
showrule(fis)
ans = 3×83 char array
'1. If (in1 is in1cluster1) and (in2 is in2cluster1) then (out1 is out1cluster1) (1)'
'2. If (in1 is in1cluster2) and (in2 is in2cluster2) then (out1 is out1cluster2) (1)'
'3. If (in1 is in1cluster3) and (in2 is in2cluster3) then (out1 is out1cluster3) (1)'
Obtain the input and output data.
load clusterDemo.dat
inputData = clusterDemo(:,1:2);
outputData = clusterDemo(:,3);Create a genfisOptions option set for FCM Clustering, specifying a Mamdani FIS type.
opt = genfisOptions("FCMClustering",FISType="mamdani");
Specify the number of clusters.
opt.NumClusters = 3;
Suppress the display of iteration information to the Command Window.
opt.Verbose = 0;
Generate the FIS.
fis = genfis(inputData,outputData,opt);
The generated FIS contains one rule for each cluster.
showrule(fis)
ans = 3×83 char array
'1. If (in1 is in1cluster1) and (in2 is in2cluster1) then (out1 is out1cluster1) (1)'
'2. If (in1 is in1cluster2) and (in2 is in2cluster2) then (out1 is out1cluster2) (1)'
'3. If (in1 is in1cluster3) and (in2 is in2cluster3) then (out1 is out1cluster3) (1)'
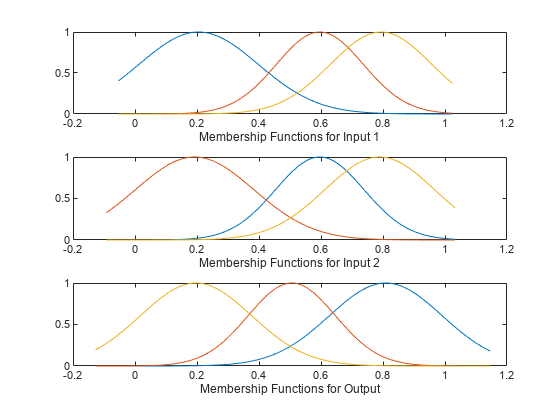
Plot the input and output membership functions.
[x,mf] = plotmf(fis,"input",1); subplot(3,1,1) plot(x,mf) xlabel("Membership Functions for Input 1") [x,mf] = plotmf(fis,"input",2); subplot(3,1,2) plot(x,mf) xlabel("Membership Functions for Input 2") [x,mf] = plotmf(fis,"output",1); subplot(3,1,3) plot(x,mf) xlabel("Membership Functions for Output")
To create a type-2 FIS from input/output data, you must first create a type-1 FIS using genfis.
Load training data and generate a FIS using subtractive clustering.
load clusterDemo.dat inputData = clusterDemo(:,1:2); outputData = clusterDemo(:,3); opt = genfisOptions('SubtractiveClustering',... 'ClusterInfluenceRange',[0.5 0.25 0.3]); fisT1 = genfis(inputData,outputData,opt); fisT1.Outputs
ans =
fisvar with properties:
Name: "out1"
Range: [-0.1274 1.1458]
MembershipFunctions: [1×3 fismf]
Convert the generated FIS to a type-2 FIS.
fisT2 = convertToType2(fisT1);
Since the initial type-1 FIS is a Sugeno system, only the input MFs are converted to type-2 MFs.
Input Arguments
Input data, specified as an N-column array, where N is the number of FIS inputs.
inputData and outputData must
have the same number of rows.
Output data, specified as an M-column array, where M is the number of FIS outputs.
When using grid partitioning, outputData must
have one column. If you specify more than one column for grid partitioning, genfis uses
the first column as the output data.
inputData and outputData must
have the same number of rows.
FIS generation options, specified as a genfisOptions option
set. If you do not specify options, genfis uses
a default grid partitioning option set.
You can generate fuzzy systems using one of the following methods, which you specify when you create the option set.
Grid partitioning — Generate input membership functions by uniformly partitioning the input variable ranges, and create a single-output Sugeno fuzzy system. The fuzzy rule base contains one rule for each input membership function combination.
options = genfisOptions("GridPartition");Subtractive clustering — Generate a Sugeno fuzzy system using membership functions and rules derived from data clusters found using subtractive clustering of input and output data. For more information on subtractive clustering, see
subclust.options = genfisOptions("SubtractiveClustering");FCM Clustering — Generate a fuzzy system using membership function and rules derived from data clusters found using FCM clustering of input and output data. For more information on FCM clustering, see
fcm.options = genfisOptions("FCMClustering");
Output Arguments
Fuzzy inference system, returned as a mamfis or
sugfis object. The properties of
fis depend on the type of clustering used and the
corresponding options.
| Clustering Type | Fuzzy System Type | Input Membership Functions | Fuzzy Rules | Output Membership Functions |
|---|---|---|---|---|
| Grid Partitioning | Sugeno | Each input variable has evenly distributed input membership functions. Specify the number of
membership functions using
options.NumMembershipFunctions.
Specify the membership function type using
options.InputMembershipFunctionType. | One rule for each input membership function combination. The consequent of each rule corresponds to a different output membership function. | One output membership function for each fuzzy rule. Specify
the membership function type using options.OutputMembershipFunctionType. |
| Subtractive Clustering | Sugeno | Each input variable has one "gaussmf" input membership function for each
fuzzy cluster. | One rule for each fuzzy cluster | Each output variable has one "linear" output membership function for each
fuzzy cluster. |
| FCM Clustering | Mamdani or Sugeno | Each input variable has one "gaussmf" input membership function for each
fuzzy cluster. | One rule for each fuzzy cluster | Each output variable has one output membership function for each fuzzy cluster. The
membership function type is "gaussmf" for
Mamdani systems and "linear" for Sugeno
systems. |
If fis is a single-output Sugeno system, you can tune the membership
function parameters using the anfis function.
Generating a type-2 FIS is not supported by genfis.
Instead, generate a type-1 FIS and convert it to a type-2 system using
convertToType2.
Alternative Functionality
App
You can interactively create a FIS from data using the Fuzzy Logic Designer app.
Version History
Introduced in R2017agenfis no longer supports fuzzy inference system
structures. Use mamfis and
sugfis
objects instead. To convert existing fuzzy inference system structures to objects,
use the convertfis function.
Support for fuzzy inference systems structures will be removed in a future
release. This change was announced in R2018b. Using fuzzy inference system
structures with genfis issues a warning starting in
R2019b.
See Also
genfisOptions | anfis | fcm | subclust
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
웹사이트 선택
번역된 콘텐츠를 보고 지역별 이벤트와 혜택을 살펴보려면 웹사이트를 선택하십시오. 현재 계신 지역에 따라 다음 웹사이트를 권장합니다:
또한 다음 목록에서 웹사이트를 선택하실 수도 있습니다.
사이트 성능 최적화 방법
최고의 사이트 성능을 위해 중국 사이트(중국어 또는 영어)를 선택하십시오. 현재 계신 지역에서는 다른 국가의 MathWorks 사이트 방문이 최적화되지 않았습니다.
미주
- América Latina (Español)
- Canada (English)
- United States (English)
유럽
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)