Specify Gradient for HMC Sampler
This example shows how to set up a Bayesian linear regression model for efficient posterior sampling using the Hamiltonian Monte Carlo (HMC) sampler. The prior distribution of the coefficients is a multivariate t, and the disturbance variance has an inverse gamma prior.
Consider the multiple linear regression model that predicts U.S. real gross national product (GNPR) using a linear combination of industrial production index (IPI), total employment (E), and real wages (WR).
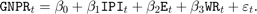
For all  ,
, 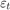 is a series of independent Gaussian disturbances with a mean of 0 and variance
is a series of independent Gaussian disturbances with a mean of 0 and variance  . Assume these prior distributions.
. Assume these prior distributions.
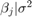 is a 4-D t distribution with 30 degrees of freedom for each component, correlation matrix
is a 4-D t distribution with 30 degrees of freedom for each component, correlation matrix C, locationct, and scalest.
 , with shape
, with shape 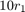 and scale
and scale  .
.
bayeslm treats these assumptions and the data likelihood as if the corresponding posterior is analytically intractable.
Declare a MATLAB® function that:
Accepts values of
 and
and  together in a column vector, and accepts values of the hyperparameters.
together in a column vector, and accepts values of the hyperparameters.Returns the log of the joint prior distribution,
 , given the values of
, given the values of  and
and  , and returns the gradient of the log prior density with respect to
, and returns the gradient of the log prior density with respect to  and
and  .
.
function [logPDF,gradient] = priorMVTIGHMC(params,ct,st,dof,C,a,b) %priorMVTIGHMC Log density of multivariate t times inverse gamma and %gradient % priorMVTIGHMC passes params(1:end-1) to the multivariate t density % function with dof degrees of freedom for each component and positive % definite correlation matrix C, and passes params(end) to the inverse % gamma distribution with shape a and scale b. priorMVTIG returns the % log of the product of the two evaluated densities and its gradient. % After applying the log, terms involving constants only are dropped from % logPDF. % % params: Parameter values at which the densities are evaluated, an % m-by-1 numeric vector. % % ct: Multivariate t distribution component centers, an (m-1)-by-1 % numeric vector. Elements correspond to the first m-1 elements % of params. % % st: Multivariate t distribution component scales, an (m-1)-by-1 % numeric vector. Elements correspond to the first m-1 elements % of params. % % dof: Degrees of freedom for the multivariate t distribution, a % numeric scalar or (m-1)-by-1 numeric vector. priorMVTIG expands % scalars such that dof = dof*ones(m-1,1). Elements of dof % correspond to the elements of params(1:end-1). % % C: correlation matrix for the multivariate t distribution, an % (m-1)-by-(m-1) symmetric, positive definite matrix. Rows and % columns correspond to the elements of params(1:end-1). % % a: Inverse gamma shape parameter, a positive numeric scalar. % % b: Inverse gamma scale parameter, a positive scalar. % beta = params(1:(end-1)); numCoeffs = numel(beta); sigma2 = params(end); tVal = (beta - ct)./st; expo = -(dof + numCoeffs)/2; logmvtDensity = expo*log(1 + (1/dof)*((tVal'/C)*tVal)); logigDensity = (-a - 1)*log(sigma2) - 1/(sigma2*b); logPDF = logmvtDensity + logigDensity; grad_beta = (expo/(1 + (1/dof)*(tVal'/C*tVal)))*((2/dof)*((tVal'/C)'./st)); grad_sigma = (-a - 1)/sigma2 + 1/(sigma2^2*b); gradient = [grad_beta; grad_sigma]; end
Create an anonymous function that operates like priorMVTIGHMC, but accepts the parameter values only and holds the hyperparameter values fixed at arbitrarily chosen values.
rng(1); % For reproducibility
dof = 30;
V = rand(4,4);
Sigma = V'*V;
st = sqrt(diag(Sigma));
C = Sigma./(st*st');
ct = -10*rand(4,1);
a = 10*rand;
b = 10*rand;
logPDF = @(params)priorMVTIGHMC(params,ct,st,dof,C,a,b);
Create a custom joint prior model for the linear regression parameters. Specify the number of predictors, p. Also, specify the function handle for priorMVTIGHMC and the variable names.
p = 3; PriorMdl = bayeslm(p,'ModelType','custom','LogPDF',logPDF,... 'VarNames',["IPI" "E" "WR"]);
PriorMdl is a customblm Bayesian linear regression model object representing the prior distribution of the regression coefficients and disturbance variance.
Load the Nelson-Plosser data set. Create variables for the response and predictor series. For numerical stability, convert the series to returns.
load Data_NelsonPlosser X = price2ret(DataTable{:,PriorMdl.VarNames(2:end)}); y = price2ret(DataTable{:,'GNPR'});
Estimate the marginal posterior distributions of  and
and  using the HMC sampler. To monitor the progress of the sampler, specify a verbosity level of
using the HMC sampler. To monitor the progress of the sampler, specify a verbosity level of 2. For numerical stability, specify reparameterization of the disturbance variance.
options = sampleroptions('Sampler','hmc','VerbosityLevel',2) PosteriorMdl = estimate(PriorMdl,X,y,'Options',options,'Reparameterize',true);
options =
struct with fields:
Sampler: 'HMC'
StepSizeTuningMethod: 'dual-averaging'
MassVectorTuningMethod: 'iterative-sampling'
NumStepSizeTuningIterations: 100
TargetAcceptanceRatio: 0.6500
NumStepsLimit: 2000
VerbosityLevel: 2
NumPrint: 100
o Tuning mass vector using method: iterative-sampling
o Initial step size for dual-averaging = 0.0625
|==================================================================================|
| ITER | LOG PDF | STEP SIZE | NUM STEPS | ACC RATIO | DIVERGENT |
|==================================================================================|
| 100 | 4.312313e+01 | 1.048e-01 | 48 | 6.300e-01 | 3 |
Finished mass vector tuning iteration 1 of 5.
o Initial step size for dual-averaging = 0.5
|==================================================================================|
| ITER | LOG PDF | STEP SIZE | NUM STEPS | ACC RATIO | DIVERGENT |
|==================================================================================|
| 100 | 4.278350e+01 | 5.691e-01 | 9 | 6.300e-01 | 0 |
|==================================================================================|
| ITER | LOG PDF | STEP SIZE | NUM STEPS | ACC RATIO | DIVERGENT |
|==================================================================================|
| 100 | 4.080779e+01 | 4.112e-02 | 5 | 8.000e-01 | 0 |
Finished mass vector tuning iteration 2 of 5.
o Initial step size for dual-averaging = 2
|==================================================================================|
| ITER | LOG PDF | STEP SIZE | NUM STEPS | ACC RATIO | DIVERGENT |
|==================================================================================|
| 100 | 4.044744e+01 | 3.088e-01 | 16 | 5.800e-01 | 0 |
|==================================================================================|
| ITER | LOG PDF | STEP SIZE | NUM STEPS | ACC RATIO | DIVERGENT |
|==================================================================================|
| 100 | 3.823729e+01 | 2.779e-01 | 4 | 7.700e-01 | 0 |
| 200 | 4.085115e+01 | 1.140e-01 | 15 | 8.433e-01 | 0 |
Finished mass vector tuning iteration 3 of 5.
o Initial step size for dual-averaging = 0.25
|==================================================================================|
| ITER | LOG PDF | STEP SIZE | NUM STEPS | ACC RATIO | DIVERGENT |
|==================================================================================|
| 100 | 4.132405e+01 | 3.157e-01 | 16 | 6.500e-01 | 0 |
|==================================================================================|
| ITER | LOG PDF | STEP SIZE | NUM STEPS | ACC RATIO | DIVERGENT |
|==================================================================================|
| 100 | 4.357148e+01 | 1.591e-02 | 2 | 8.050e-01 | 0 |
| 200 | 3.890737e+01 | 2.841e-01 | 16 | 8.533e-01 | 0 |
Finished mass vector tuning iteration 4 of 5.
o Initial step size for dual-averaging = 0.25
|==================================================================================|
| ITER | LOG PDF | STEP SIZE | NUM STEPS | ACC RATIO | DIVERGENT |
|==================================================================================|
| 100 | 4.358423e+01 | 3.442e-01 | 15 | 6.700e-01 | 0 |
|==================================================================================|
| ITER | LOG PDF | STEP SIZE | NUM STEPS | ACC RATIO | DIVERGENT |
|==================================================================================|
| 100 | 4.048687e+01 | 9.933e-02 | 4 | 7.950e-01 | 0 |
| 200 | 4.334420e+01 | 3.098e-01 | 4 | 8.433e-01 | 0 |
Finished mass vector tuning iteration 5 of 5.
o Tuning step size using method: dual-averaging. Target acceptance ratio = 0.65
o Initial step size for dual-averaging = 0.5
|==================================================================================|
| ITER | LOG PDF | STEP SIZE | NUM STEPS | ACC RATIO | DIVERGENT |
|==================================================================================|
| 100 | 4.302411e+01 | 2.995e-01 | 17 | 6.100e-01 | 0 |
Method: MCMC sampling with 10000 draws
Number of observations: 61
Number of predictors: 4
| Mean Std CI95 Positive Distribution
-----------------------------------------------------------------------
Intercept | 0.0077 0.0086 [-0.009, 0.024] 0.812 Empirical
IPI | -0.1632 0.1461 [-0.476, 0.099] 0.123 Empirical
E | 2.3694 0.5464 [ 1.396, 3.564] 1.000 Empirical
WR | -0.2309 0.3023 [-0.844, 0.338] 0.219 Empirical
Sigma2 | 0.0036 0.0007 [ 0.002, 0.005] 1.000 Empirical
PosteriorMdl is an empiricalblm model object storing the draws from the posterior distributions.
View trace plots and autocorrelation function (ACF) plots of the draws from the posteriors.
for j = 1:4 figure; subplot(2,1,1) plot(PosteriorMdl.BetaDraws(j,:)); title(sprintf('Trace plot - Beta %d',j)); xlabel('MCMC draw') ylabel('Simulation index') subplot(2,1,2) autocorr(PosteriorMdl.BetaDraws(j,:)) end figure; subplot(2,1,1) plot(PosteriorMdl.Sigma2Draws); title('Trace plot - Disturbance Variance'); xlabel('MCMC draw') ylabel('Simulation index') subplot(2,1,2) autocorr(PosteriorMdl.Sigma2Draws)
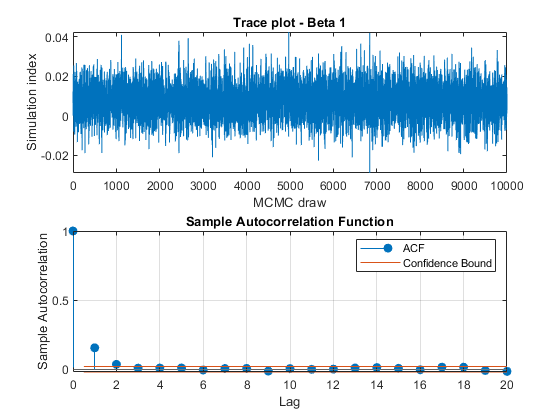

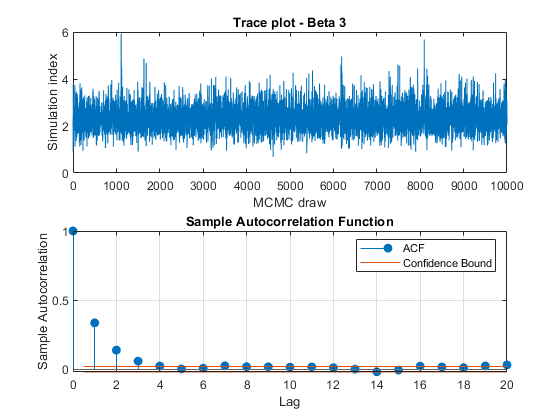
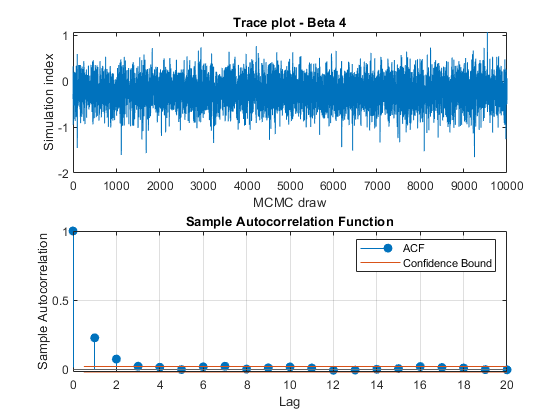
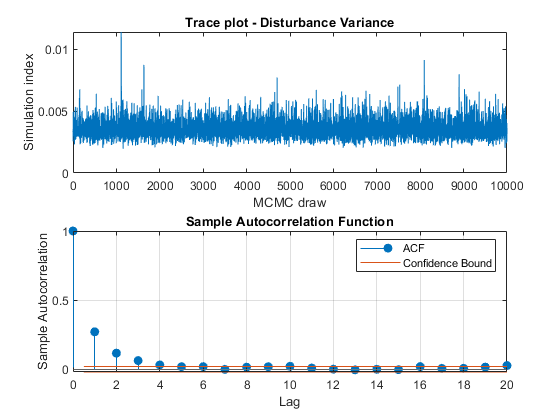
The trace plots show adequate mixing and no transient behavior. The autocorrelation plots show low correlation among subsequent samples.