gcrmabackadj
Perform GC Robust Multi-array Average (GCRMA) background adjustment on Affymetrix microarray probe-level data using sequence information
Syntax
PMMatrix_Adj = gcrmabackadj(PMMatrix, MMMatrix, AffinPM, AffinMM)
[PMMatrix_Adj, nsbStruct]
= gcrmabackadj(PMMatrix, MMMatrix, AffinPM, AffinMM)
... = gcrmabackadj( ...'OpticalCorr', OpticalCorrValue,
...)
... = gcrmabackadj( ...'CorrConst', CorrConstValue,
...)
... = gcrmabackadj( ...'Method', MethodValue,
...)
... = gcrmabackadj( ...'TuningParam', TuningParamValue,
...)
... = gcrmabackadj( ...'AddVariance', AddVarianceValue,
...)
... = gcrmabackadj( ...'GSBCorr', GSBCorrValue,
...)
... = gcrmabackadj( ...'Showplot', ShowplotValue,
...)
... = gcrmabackadj( ...'Verbose', VerboseValue,
...)
Input Arguments
PMMatrix | Matrix of intensity values where each row corresponds to a perfect match (PM) probe and each column corresponds to an Affymetrix® CEL file. (Each CEL file is generated from a separate chip. All chips should be of the same type.) Tip You can use the
|
MMMatrix | Matrix of intensity values where each row corresponds to a mismatch (MM) probe and each column corresponds to an Affymetrix CEL file. (Each CEL file is generated from a separate chip. All chips should be of the same type.) Tip You can use the
|
AffinPM | Column vector of PM probe affinities, such as returned by the affyprobeaffinities function. Each row
corresponds to a probe. |
AffinMM | Column vector of MM probe affinities, such as returned by the affyprobeaffinities function. Each row
corresponds to a probe. |
OpticalCorrValue | Controls the use of optical background correction on the PM
and MM probe intensity values in PMMatrix and MMMatrix.
Choices are true (default) or false. |
CorrConstValue | Value that specifies the correlation constant, rho, for log
background intensity for each PM/MM probe pair. Choices are any value ≥
0 and ≤ 1. Default is 0.7. |
MethodValue | Character vector or string that specifies the method to estimate the signal. Choices are
'MLE', a faster, ad hoc Maximum
Likelihood Estimate method, or
'EB', a slower, more formal,
empirical Bayes method. Default is
'MLE'. |
TuningParamValue | Value that specifies the tuning parameter used by the estimate
method. This tuning parameter sets the lower bound of signal values
with positive probability. Choices are a positive value. Default is 5 (MLE)
or 0.5 (EB).Tip For information on determining a setting for this parameter, see Wu et al., 2004. |
AddVarianceValue | Controls whether the signal variance is added to the weight
function for smoothing low signal edge. Choices are true or false (default). |
GSBCorrValue | Specifies whether to perform gene-specific binding (GSB) correction
using probe affinity data. Choices are true (default)
or false. If there is no probe affinity information,
this property is ignored. |
ShowplotValue | Controls the display of a plot showing the log2 of
probe intensity values from a specified column (chip) in MMMatrix,
versus probe affinities in AffinMM. Choices
are true, false, or I,
an integer specifying a column in MMMatrix.
If set to true, the first column in MMMatrix is
plotted. Default is:
|
VerboseValue | Controls the display of a progress report showing the number
of each chip as it is completed. Choices are true (default)
or false. |
Output Arguments
PMMatrix_Adj | Matrix of background adjusted PM (perfect match) intensity values. |
nsbStruct | Structure containing nonspecific binding background parameters,
estimated from the intensities and affinities of probes on an Affymetrix GeneChip® array.
|
Description
PMMatrix_Adj = gcrmabackadj(PMMatrix, MMMatrix, AffinPM, AffinMM)PMMatrix_Adj,
a matrix of background adjusted PM (perfect match) intensity values.
Note
If AffinPM and AffinMM data
are not available, you can still use the gcrmabackadj function
by entering empty column vectors for both of these inputs in the syntax.
[ returns PMMatrix_Adj, nsbStruct]
= gcrmabackadj(PMMatrix, MMMatrix, AffinPM, AffinMM)nsbStruct,
a structure containing nonspecific binding background parameters,
estimated from the intensities and affinities of probes on an Affymetrix GeneChip array. nsbStruct includes
the following fields:
sigmamu_pmmu_mm
... = gcrmabackadj( ...' calls PropertyName', PropertyValue,
...)gcrmabackadj with optional
properties that use property name/property value pairs. You can specify
one or more properties in any order. Each PropertyName must
be enclosed in single quotation marks and is case insensitive. These
property name/property value pairs are as follows:
... = gcrmabackadj( ...'OpticalCorr', controls the use of optical background correction
on the PM and MM probe intensity values in OpticalCorrValue,
...)PMMatrix and MMMatrix.
Choices are true (default) or false.
... = gcrmabackadj( ...'CorrConst', specifies the correlation constant, rho, for log background
intensity for each PM/MM probe pair. Choices are any value CorrConstValue,
...)≥
0 and ≤ 1. Default is 0.7.
... = gcrmabackadj( ...'Method', specifies the method to estimate the signal. Choices
are MethodValue,
...)MLE, a faster, ad hoc Maximum Likelihood Estimate
method, or EB, a slower, more formal, empirical
Bayes method. Default is MLE.
... = gcrmabackadj( ...'TuningParam', specifies the tuning parameter used by the estimate
method. This tuning parameter sets the lower bound of signal values
with positive probability. Choices are a positive value. Default is TuningParamValue,
...)5 (MLE)
or 0.5 (EB).
Tip
For information on determining a setting for this parameter, see Wu et al., 2004.
... = gcrmabackadj( ...'AddVariance', controls whether the signal variance is added to the
weight function for smoothing low signal edge. Choices are AddVarianceValue,
...)true or false (default).
... = gcrmabackadj( ...'GSBCorr', specifies whether to perform gene specific binding
(GSB) correction using probe affinity data. Choices are GSBCorrValue,
...)true (default)
or false. If there is no probe affinity information,
this property is ignored.
... = gcrmabackadj( ...'Showplot', controls the display of a plot showing the log2 of
probe intensity values from a specified column (chip) in ShowplotValue,
...)MMMatrix,
versus probe affinities in AffinMM. Choices
are true, false, or I,
an integer specifying a column in MMMatrix.
If set to true, the first column in MMMatrix is
plotted. Default is:
false— When return values are specified.true— When return values are not specified.
... = gcrmabackadj( ...'Verbose', controls the display of a progress report showing
the number of each chip as it is completed. Choices are VerboseValue,
...)true (default)
or false.
Examples
Load the MAT-file, included with the Bioinformatics Toolbox™ software, that contains Affymetrix data from a prostate cancer study. The variables in the MAT-file include
seqMatrix, a matrix containing sequence information for PM probes,pmMatrixandmmMatrix, matrices containing PM and MM probe intensity values, andprobeIndices, a column vector containing probe indexing information.load prostatecancerrawdataCompute the Affymetrix PM and MM probe affinities from their sequences and MM probe intensities.
[apm, amm] = affyprobeaffinities(seqMatrix, mmMatrix(:,1),... 'ProbeIndices', probeIndices);
Perform GCRMA background adjustment on the Affymetrix microarray probe-level data, creating a matrix of background adjusted PM intensity values. Also, display a plot showing the log2 of probe intensity values from column 3 (chip 3) in
mmMatrix, versus probe affinities inamm.pms_adj = gcrmabackadj(pmMatrix, mmMatrix, apm, amm, 'showplot', 3);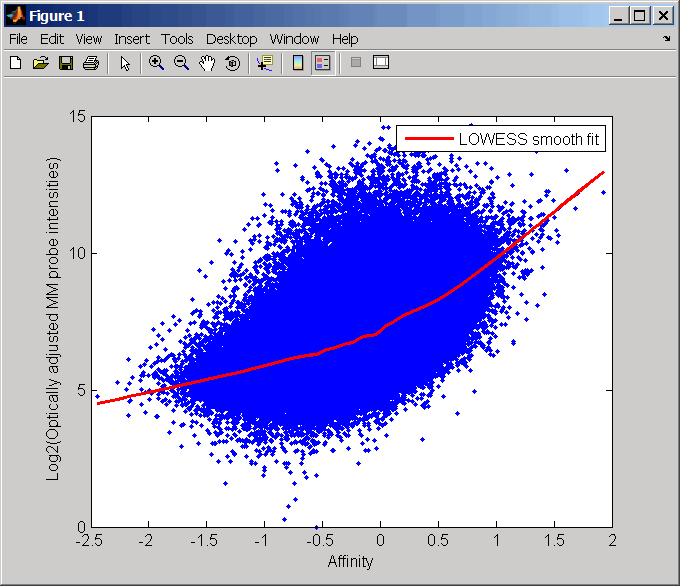
Perform GCRMA background adjustment again, using the slower, more formal, empirical Bayes method.
pms_adj2 = gcrmabackadj(pmMatrix, mmMatrix, apm, amm, 'method', 'EB');
The prostatecancerrawdata.mat file used in
this example contains data from Best et al., 2005.
References
[1] Wu, Z., Irizarry, R.A., Gentleman, R., Murillo, F.M., and Spencer, F. (2004). A Model Based Background Adjustment for Oligonucleotide Expression Arrays. Journal of the American Statistical Association 99(468), 909–917.
[2] Wu, Z., and Irizarry, R.A. (2005). Stochastic Models Inspired by Hybridization Theory for Short Oligonucleotide Arrays. Proceedings of RECOMB 2004. J Comput Biol. 12(6), 882–93.
[3] Wu, Z., and Irizarry, R.A. (2005). A Statistical Framework for the Analysis of Microarray Probe-Level Data. Johns Hopkins University, Biostatistics Working Papers 73.
[4] Wu, Z., and Irizarry, R.A. (2003). A Model Based Background
Adjustment for Oligonucleotide Expression Arrays.
RSS Workshop on Gene Expression, Wye, England,
https://biosun01.biostat.jhsph.edu/%7Eririzarr/Talks/gctalk.pdf.
[5] Abd Rabbo, N.A., and Barakat, H.M. (1979). Estimation Problems in Bivariate Lognormal Distribution. Indian J. Pure Appl. Math 10(7), 815–825.
[6] Best, C.J.M., Gillespie, J.W., Yi, Y., Chandramouli, G.V.R., Perlmutter, M.A., Gathright, Y., Erickson, H.S., Georgevich, L., Tangrea, M.A., Duray, P.H., Gonzalez, S., Velasco, A., Linehan, W.M., Matusik, R.J., Price, D.K., Figg, W.D., Emmert-Buck, M.R., and Chuaqui, R.F. (2005). Molecular alterations in primary prostate cancer after androgen ablation therapy. Clinical Cancer Research 11, 6823–6834.
Version History
Introduced in R2007a
See Also
affygcrma | affyprobeseqread | affyread | celintensityread | probelibraryinfo
